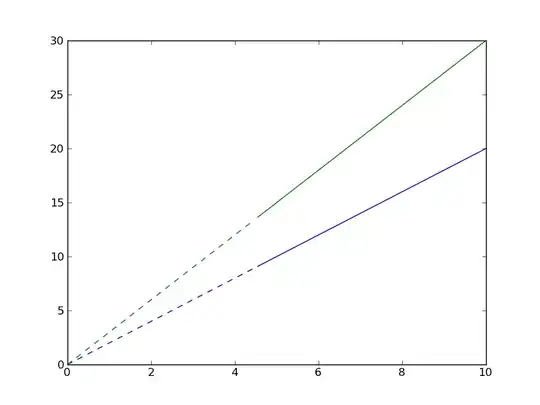
Edit: I'd had this open and left, so I didn't notice @Ricardo's answer. Because matplotlib will convert things to numpy arrays regardless, there are more efficient ways to do it.
As an example:
Just plot two different lines, one with a dashed linestyle and another with a solid linestyle.
E.g.
import numpy as np
import matplotlib.pyplot as plt
x = np.linspace(0, 10, 100)
y1 = 2 * x
y2 = 3 * x
xthresh = 4.5
diff = np.abs(y1 - y2)
below = diff < xthresh
above = diff >= xthresh
# Plot lines below threshold as dotted...
plt.plot(x[below], y1[below], 'b--')
plt.plot(x[below], y2[below], 'g--')
# Plot lines above threshold as solid...
plt.plot(x[above], y1[above], 'b-')
plt.plot(x[above], y2[above], 'g-')
plt.show()
For the case where they're cyclic, use masked arrays:
import numpy as np
import matplotlib.pyplot as plt
x = np.linspace(0, 10, 100)
y1 = 2 * np.cos(x)
y2 = 3 * np.sin(x)
xthresh = 2.0
diff = np.abs(y1 - y2)
below = diff < xthresh
above = diff >= xthresh
# Plot lines below threshold as dotted...
plt.plot(np.ma.masked_where(below, x), np.ma.masked_where(below, y1), 'b--')
plt.plot(np.ma.masked_where(below, x), np.ma.masked_where(below, y2), 'g--')
# Plot lines above threshold as solid...
plt.plot(np.ma.masked_where(above, x), np.ma.masked_where(above, y1), 'b-')
plt.plot(np.ma.masked_where(above, x), np.ma.masked_where(above, y2), 'g-')
plt.show()