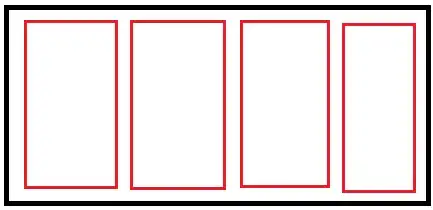
From a CFD simulation in Ansys Fluent, I have a dataframe from a csv containing 4 columns: x,y,z (nodal coordinates of irregular grid), and u (streamwise velocity magnitude). I want to plot a contour plot of u over the x-z plane. The solid body around which air flows has u=0, and I want this to be masked for the interpolation, i.e. I want to create a contour plot with an irregular boundary. The first attached image is more or less what I'd like to get with Python, obtained with Ansys CFD-Post -- the geometry of the solid body is outlined in black:
This is what the data looks like. Note column values are not unique or sorted.
### geometric parameters, in m
L = 0.3048
hwall = 0.0095
Lin = L-hwall*2
H = 0.073
Lpad = Lin/10
Hpad = H/10
### get df: x,y,z coordinates and instantaneous streamwise velocity
df = pd.read_csv('test2.csv',names=['x','y','z','u'])
dfsub = df.loc[(df.x>-Lin/2-Lpad)&(df.x<Lin/2+Lpad)&(df.z<H+Hpad)] # limit to smaller region
dfsub
x y z u
2 -0.141747 2.106994e-11 0.010645 0.106242
3 -0.139540 -6.458060e-12 0.010615 0.456511
5 -0.132303 -1.308423e-12 0.010602 1.138072
8 0.141747 -2.106730e-11 0.010650 -0.154759
9 0.139543 1.165080e-11 0.010619 -0.150316
... ... ... ... ...
3597 0.159718 -2.567698e-11 0.027614 -0.532316
3598 0.159934 2.542068e-13 0.027258 -0.544210
3599 0.159501 1.936669e-12 0.027613 -0.520241
3600 0.157081 -6.854833e-12 0.035597 -0.392042
3601 0.156974 1.765435e-11 0.027732 -0.382951
Here is a scatterplot to show the data:
umax = max(np.abs(dfsub.u))
fig,ax=plt.subplots(figsize=(16,3))
dfsub.plot.scatter('x','z',c='u',cmap ='seismic',vmin=-umax,vmax=umax,ax=ax)
plt.show()
And here is the basic code for plotting, which I don't expect to work since the solid region is just read as velocity = 0 for interpolation:
### contour plot
crange = 4
fig,ax=plt.subplots(figsize=(16,3))
tcf = ax.tricontourf(dfsub.x,dfsub.z,dfsub.u,levels=np.arange(
crange,crange,0.1),cmap='seismic',extend='both')
plt.colorbar(tcf,ax=ax,label='$u$ (m/s)')
ax.set_title('basic',size=14)
plt.show()
and here is the output:
I've tried to mask these values in several ways, e.g. replace 0s with nans, which gives "ValueError: z array must not contain non-finite values within the triangulation":
### get df: x,y,z coordinates and instantaneous streamwise velocity
df = pd.read_csv('test2.csv',names=['x','y','z','u'])
df.replace(0, np.nan, inplace=True)
dfsub = df.loc[(df.x>-Lin/2-Lpad)&(df.x<Lin/2+Lpad)&(df.z<H+Hpad)] # limit to smaller region
or to remove the 0s from the dataframe:
### get df: x,y,z coordinates and instantaneous streamwise velocity
df = pd.read_csv('test2.csv',names=['x','y','z','u'])
df = df.loc[df.u != 0]
dfsub = df.loc[(df.x>-Lin/2-Lpad)&(df.x<Lin/2+Lpad)&(df.z<H+Hpad)] # limit to smaller region
The output for this is in the attached image. It still interpolates in the regions I mean to exclude.