It appears that for larger nnet::multinom multinomial regression models (with a few thousand coefficients), calculating the Hessian (the matrix of second derivatives of the negative log likelihood, also known as the observed Fisher information matrix) becomes super slow, which then prevents me from calculating the variance-covariance matrix & allowing me to calculate confidence intervals on model predictions.
It seems the culprit is the following pure R function - it seems it uses some code to calculate the Fisher information matrix analytically using code contributed by David Firth : https://github.com/cran/nnet/blob/master/R/vcovmultinom.R
multinomHess = function (object, Z = model.matrix(object))
{
probs <- object$fitted
coefs <- coef(object)
if (is.vector(coefs)) {
coefs <- t(as.matrix(coefs))
probs <- cbind(1 - probs, probs)
}
coefdim <- dim(coefs)
p <- coefdim[2L]
k <- coefdim[1L]
ncoefs <- k * p
kpees <- rep(p, k)
n <- dim(Z)[1L]
## Now compute the observed (= expected, in this case) information,
## e.g. as in T Amemiya "Advanced Econometrics" (1985) pp 295-6.
## Here i and j are as in Amemiya, and x, xbar are vectors
## specific to (i,j) and to i respectively.
info <- matrix(0, ncoefs, ncoefs)
Names <- dimnames(coefs)
if (is.null(Names[[1L]]))
Names <- Names[[2L]]
else Names <- as.vector(outer(Names[[2L]], Names[[1L]], function(name2,
name1) paste(name1, name2, sep = ":")))
dimnames(info) <- list(Names, Names)
x0 <- matrix(0, p, k + 1L)
row.totals <- object$weights
for (i in seq_len(n)) {
Zi <- Z[i, ]
xbar <- rep(Zi, times=k) * rep(probs[i, -1, drop=FALSE], times=kpees)
for (j in seq_len(k + 1)) {
x <- x0
x[, j] <- Zi
x <- x[, -1, drop = FALSE]
x <- x - xbar
dim(x) <- c(1, ncoefs)
info <- info + (row.totals[i] * probs[i, j] * crossprod(x))
}
}
info
}
The info in the Advanced Econometrics book that is referenced states
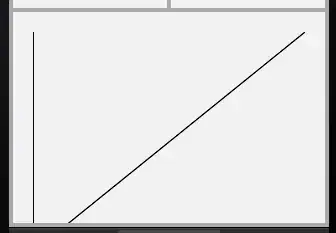
From this explanation, we can see that the Hessian indeed is just given by the sum of a bunch of crossproducts. I also saw this and this in terms of derivation of how to calculate the Hessian matrix of a multinomial regression model, which may be even more elegant and efficient, as the Hessian is there calculated based on a sum of Kronecker products.
For a smallish nnet::multinom model (in which I am modelling the frequency of different SARS-CoV2 lineages through time) the provided function runs quickly :
library(nnet)
library(splines)
download.file("https://www.dropbox.com/s/gt0yennn2gkg3rd/smallmodel.RData?dl=1",
"smallmodel.RData",
method = "auto", mode="wb")
load("smallmodel.RData")
length(fit_multinom_small$lev) # k=12 outcome levels
dim(coef(fit_multinom_small)) # 11 x 3 = (k-1) x p = 33 coefs
system.time(hess <- nnet:::multinomHess(fit_multinom_small)) # 0.11s
dim(hess) # 33 33
but doing this for a large model takes more than 2 hours (even though the model itself fits in ca. 1 minute) (again modelling the frequency of different SARS-CoV2 lineages through time, but now across different continents / countries) :
download.file("https://www.dropbox.com/s/mpz08jj7fmubd68/bigmodel.RData?dl=1",
"bigmodel.RData",
method = "auto", mode="wb")
load("bigmodel.RData")
length(fit_global_multi_last3m$lev) # k=20 outcome levels
dim(coef(fit_global_multi_last3m)) # 19 x 229 = (k-1) x p = 4351 coefficients
system.time(hess <- nnet:::multinomHess(fit_global_multi_last3m)) # takes forever
I was now looking for ways to speed up the above function.
The obvious attempt could be to port it to Rcpp, but unfortunately I am not so experienced in this. Anybody any thoughts?
EDIT: From the info here and here, it appears that calculating the Hessian for a multinomial fit should just come down to calculating a sum of Kronecker products, which we can just do from R using efficient matrix algebra, but right now I am unsure how to include my total row counts fit$weights. Anybody any idea?
download.file("https://www.dropbox.com/s/gt0yennn2gkg3rd/smallmodel.RData?dl=1",
"smallmodel.RData",
method = "auto", mode="wb")
load("smallmodel.RData")
library(nnet)
length(fit_multinom_small$lev) # k=12 outcome levels
dim(coef(fit_multinom_small)) # 11 x 3 = (k-1) x p = 33 coefs
fit = fit_multinom_small
Z = model.matrix(fit)
P = fitted(fit)[, -1, drop=F]
k = ncol(P) # nr of outcome categories-1
p = ncol(Z) # nr of parameters
n = nrow(Z) # nr of observations
ncoefs = k*p
library(fastmatrix)
# Fisher information matrix
info <- matrix(0, ncoefs, ncoefs)
for (i in 1:n) { # sum over observations
info = info + kronecker.prod(diag(P[i,]) - tcrossprod(P[i,]), tcrossprod(Z[i,]))
}