I'm trying to come up with a way to use Keras-Tuner to auto-identify the best parameters for my CNN. I am using Celeb_a dataset
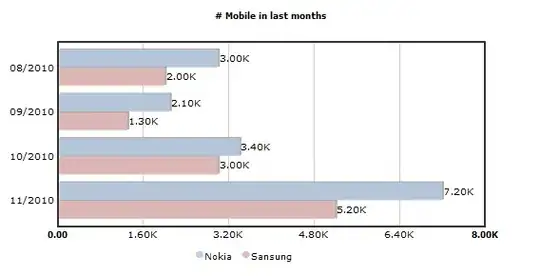
I tried a similar project where I used fashion_mnist and this worked perfectly but my experience with python isn't enough to do what I want to achieve. When I tried with fashion_mnist I managed to create this table of results
My code is here.
I am hoping to produce a similar table using the Celeb_a dataset. This is for a report I'm doing for college. In the report, my college used AWS Rekognition to produce the table below.
I am hoping to be able to train the data so I can save this model to a pickle and generate a similar table of results to compare them.
Any recommendations on how to approach this? My queries at the moment are:
- How to load the dataset correctly?
- how can i train the model to give me accuracy on "Moustache", "Beard", "Emotion" (like on the table of results above)
I tried loading the data using:
(x_train, y_train), (x_test, y_test) = tfds.load('celeb_a')
but this gives me the following error
AttributeError: Failed to construct dataset celeb_a: module 'tensorflow_datasets.core.utils' has no attribute 'version'
I am using:
Conda: TensorFlow (Python 3.8.5)
Windows 10 Pro
Intel(R) Core(TM) i3-4170 CPU @ 3.7GHz
64-bit
This is the script I am using to start, the same as the one in my bitbucket, Any help would be appreciated. Thank you in advance.
# -*- coding: utf-8 -*-
import tensorflow_datasets as tfds
#from tensorflow.keras.datasets import fashion_mnist
#import matplotlib.pyplot as plt
from tensorflow import keras
from tensorflow.keras.layers import Conv2D, MaxPooling2D, Dense, Flatten, Activation
from kerastuner.tuners import RandomSearch
#from kerastuner.engine.hyperparameters import HyperParameter
import time
import os
LOG_DIR = f"{int(time.time())}"
(x_train, y_train), (x_test, y_test) = tfds.load('celeb_a')
x_train = x_train.reshape(-1,28,28,1)
x_test = x_test.reshape(-1,28,28,1)
def build_model(hp): #random search passes this hyperparameter() object
model = keras.models.Sequential()
#model.add(Conv2D(32, (3, 3), input_shape=x_train.shape[1:]))
model.add(Conv2D(hp.Int("input_units", min_value=32, max_value=256, step=32), (3,3), input_shape = x_train.shape[1:]))
model.add(Activation('relu'))
model.add(MaxPooling2D(pool_size=(2, 2)))
for i in range(hp.Int("n_layers",min_value = 1, max_value = 4, step=1)):
#model.add(Conv2D(32, (3, 3)))
model.add(Conv2D(hp.Int(f"conv_{i}_units", min_value=32, max_value=256, step=32), (3,3)))
model.add(Activation('relu'))
#model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Flatten()) # this converts our 3D feature maps to 1D feature vectors
model.add(Dense(10))
model.add(Activation("softmax"))
model.compile(optimizer="adam",
loss="sparse_categorical_crossentropy",
metrics=["accuracy"])
return model
tuner = RandomSearch(build_model,
objective = "val_accuracy",
max_trials = 1,
executions_per_trial=1, #BEST PERFOMANCE SET TO 3+
directory= os.path.normpath('C:/'),# there is a limit of characters keep path short
overwrite=True #need this to override model when testing
)
tuner.search(x=x_train,
y=y_train,
epochs=1,
batch_size=64,
validation_data=(x_test,y_test),)