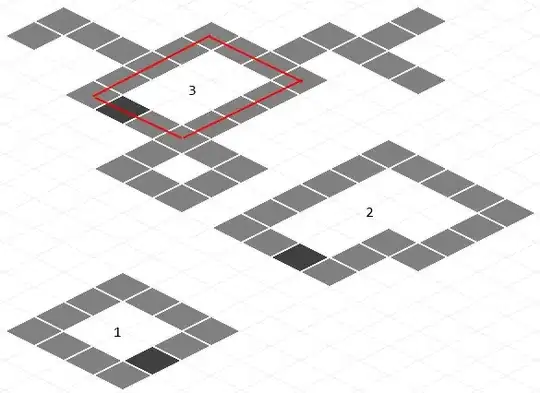
You can use partition for this:
partition splits an element set into equivalency classes. You can define your equivalence class as all points within a given euclidean distance (radius tolerance)
If you have C++11, you can simply use a lambda function:
int th_distance = 18; // radius tolerance
int th2 = th_distance * th_distance; // squared radius tolerance
vector<int> labels;
int n_labels = partition(pts, labels, [th2](const Point& lhs, const Point& rhs) {
return ((lhs.x - rhs.x)*(lhs.x - rhs.x) + (lhs.y - rhs.y)*(lhs.y - rhs.y)) < th2;
});
otherwise, you can just build a functor (see details in the code below).
With appropriate radius distance (I found 18 works good on this image), I got:
Full code:
#include <opencv2\opencv.hpp>
#include <vector>
#include <algorithm>
using namespace std;
using namespace cv;
struct EuclideanDistanceFunctor
{
int _dist2;
EuclideanDistanceFunctor(int dist) : _dist2(dist*dist) {}
bool operator()(const Point& lhs, const Point& rhs) const
{
return ((lhs.x - rhs.x)*(lhs.x - rhs.x) + (lhs.y - rhs.y)*(lhs.y - rhs.y)) < _dist2;
}
};
int main()
{
// Load the image (grayscale)
Mat1b img = imread("path_to_image", IMREAD_GRAYSCALE);
// Get all non black points
vector<Point> pts;
findNonZero(img, pts);
// Define the radius tolerance
int th_distance = 18; // radius tolerance
// Apply partition
// All pixels within the radius tolerance distance will belong to the same class (same label)
vector<int> labels;
// With functor
//int n_labels = partition(pts, labels, EuclideanDistanceFunctor(th_distance));
// With lambda function (require C++11)
int th2 = th_distance * th_distance;
int n_labels = partition(pts, labels, [th2](const Point& lhs, const Point& rhs) {
return ((lhs.x - rhs.x)*(lhs.x - rhs.x) + (lhs.y - rhs.y)*(lhs.y - rhs.y)) < th2;
});
// You can save all points in the same class in a vector (one for each class), just like findContours
vector<vector<Point>> contours(n_labels);
for (int i = 0; i < pts.size(); ++i)
{
contours[labels[i]].push_back(pts[i]);
}
// Draw results
// Build a vector of random color, one for each class (label)
vector<Vec3b> colors;
for (int i = 0; i < n_labels; ++i)
{
colors.push_back(Vec3b(rand() & 255, rand() & 255, rand() & 255));
}
// Draw the labels
Mat3b lbl(img.rows, img.cols, Vec3b(0, 0, 0));
for (int i = 0; i < pts.size(); ++i)
{
lbl(pts[i]) = colors[labels[i]];
}
imshow("Labels", lbl);
waitKey();
return 0;
}