I have found that when I try to overlay multiple rasters using plot(...,add=T) if I try to overlay more than 3 rasters together the subsequent plot does not align the rasters properly.
My original intent was to create a categorical map of modeled landcover where the darkness of the color representing a cover class varied wrt the certainty in our model projection. To do this, I created a simple script that would loop through each cover class and plot it (e.g., forest, green color on map) using a color gradient from grey (low certainty forest prediction) to full cover color (e.g., dark green for areas are strongly predicted). What I have found is that using this approach, after the 3rd cover is added to the plot, all subsequent rasters that are overlayed on the plot are arbitrarily misaligned. I have reversed the order of plotting of the cover classes and the same behavior is exhibited meaning it is not an issue with the individual cover class rasters. Even more puzzling in Rstudio, when I use the zoom button to closely inspect the final plot, the misalignment worsens.
Do you have any ideas of why this behavior exists? Most importantly, do you have any suggested solutions or workarounds?
The code and data on the link below has all of the behaviors described captured. https://dl.dropboxusercontent.com/u/332961/r%20plot%20raster%20add%20issue.zip Turn plot_gradient=F to see how if you just simply subset a same raster and add the subsets sequentially to the same plot you can replicate the issue. I have already tried setting the extent of the plot device plot(..., ext) but that did not work. I have also checked and the extent of each cover raster is the same.
Below is the figure of the misaligned cover classes. plotting to jpeg device will result in a similar image (i.e., this is not an issue of Rstudio rendering).
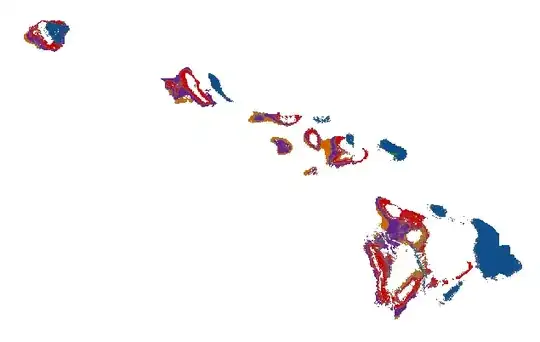 Strangely enough, if I zoom into the image using Rstudio, the misalignment is different
Strangely enough, if I zoom into the image using Rstudio, the misalignment is different
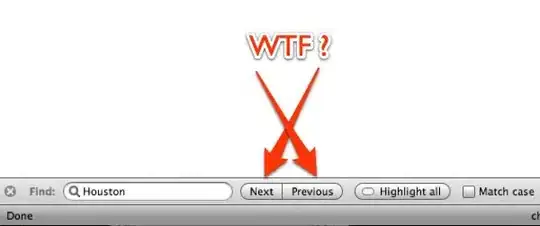 For comparison, this is how the covers should align correctly in the landscape
For comparison, this is how the covers should align correctly in the landscape
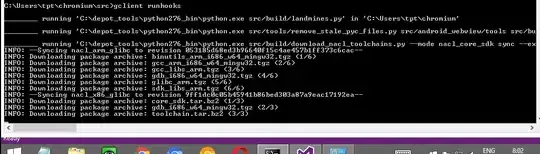
library(raster)
library(colorRamps)
raster_of_classes=raster("C:/r plot raster add issue/raster_of_classes.tif")
raster_of_certainty_of_classes=raster("C:/r plot raster add issue/raster_of_certainty_of_classes.tif")
endCols=c("darkorchid4", "darkorange3", "red3", "green4", "dodgerblue4") #colors to be used in gradients for each class
classes=unique(raster_of_classes)
minVal=cellStats(raster_of_certainty_of_classes, min)
tmp_i=1
addPlot=F
plot_gradient=F #this is for debug only
#classes=rev(classes) #turn this off and on to see how last 2 classes are mis aligned, regardless of plotting order
for (class in classes){
raster_class=raster_of_classes==class #create mask for individual class
raster_class[raster_class==0]=NA #remove 0s from mask so they to do not get plotted
if (plot_gradient){
raster_of_certainty_of_class=raster_of_certainty_of_classes*raster_class #apply class mask to certainty map
}else{
raster_of_certainty_of_class=raster_class #apply class mask to certainty map
}
endCol=endCols[tmp_i] #pick color for gradient
col5 <- colorRampPalette(c('grey50', endCol))
if (plot_gradient){
plot(raster_of_certainty_of_class,
col=col5(n=49), breaks=seq(minVal,1,length.out=50), #as uncertainty values range from 0 to 1 plot them with fixed range
useRaster=T, axes=FALSE, box=FALSE, add=addPlot, legend=F)
}else{
plot(raster_of_certainty_of_class,
col=endCol,
useRaster=T, axes=FALSE, box=FALSE, add=addPlot, legend=F)
}
tmp_i=tmp_i+1
addPlot=T #after plotting first class, all other classes are added
}