I have an image (200x200) and want to find the neighborhood locations in a specific point with a predefined radius. For example, with the radius of 5, I have 25 points around a point. Does MATLAB can do it? The problem is about the edge of image which it does not always 25 points and the program should just find the points that are within that radius. These points can be varied from 1 (corner) to 25 (center of image)
Asked
Active
Viewed 3,033 times
3
-
related questions: [MATLAB: how do I crop out a circle from an image](http://stackoverflow.com/q/4651778/97160), [MATLAB/Octave: cut a lot of circles from a image](http://stackoverflow.com/q/7584324/97160) – Amro Aug 08 '12 at 01:32
2 Answers
6
Here is an example:
%# sample grayscale image
img = imread('cameraman.tif');
[imgH,imgW,~] = size(img);
%# circle params
t = linspace(0, 2*pi, 50); %# approximate circle with 50 points
r = 80; %# radius
c = [100 130]; %# center
%# get circular mask
BW = poly2mask(r*cos(t)+c(1), r*sin(t)+c(2), imgH, imgW);
%# show cropped image
imshow( immultiply(img,BW) )
axis on
This will handle edges cases just fine. The advantage of using POLY2MASK is that it computes the mask with a sub-pixel accuracy (read the algorithm section in the function documentation), provided you are using enough points to approximate the circle.
Amro
- 123,847
- 25
- 243
- 454
-
if you dont have access to the image processing toolbox, you could do something similar using MESHGRID/NDGRID – Amro Aug 08 '12 at 01:23
-
Thanks a lot Amro. Just two questions: 1)How I can change the circle to square and 2) If I have a 3D matrix, then what should I do? – Sam Aug 08 '12 at 02:15
-
@Sam: 1) a square mask is simple: `mask = false(imgH,imgW); mask(100:200,100:200) = true;` 2) if you have an RGB image, you could simply replicate the mask along the third dimension `repmat(mask,[1 1 3])` (assuming all channels are masked the same, otherwise process each R,G,B channel separately then combine back with `cat(3,R,G,B)`) – Amro Aug 08 '12 at 02:35
-
@ Amro: Actually I have a 3D matrix (100x100x50) and I want to find the location (or indices) of neighborhood of a point on that matrix. For example, if I consider a cubic as the desired neighborhood, then in the center I can say that my neighborhood are (i-size_of_cubic/2:i+size_of_cubic,j-size_of_cubic/2:j+size_of_cubic,k-size_of_cubic/2:k+size_of_cubic). I need a function that can do it for me and like your circle function can encounter with corners (in the corners I have just a quarter of cubic. Thanks for your help – Sam Aug 08 '12 at 03:33
-
@Sam: since the question was tagged as image-processing, I assumed you were working with images. Also the "radius" got to me to think you wanted a circular neighborhood, but now I see that's not what you meant (an illustration would have been helpful)... Anyway, check out the following two answers, I think they should help (one is for the 2D case, the other for the 3D case): [MATLAB moving a point in the XY plane](http://stackoverflow.com/a/11133312/97160), [3 dimensional matrices](http://stackoverflow.com/a/6624411/97160). – Amro Aug 08 '12 at 16:07
-
@Sam: I [added](http://stackoverflow.com/a/11869406/97160) another solution that should answer your question (its based on the one I just mentioned). I am keeping the answer above, as it might be helpful for others.. – Amro Aug 08 '12 at 16:54
2
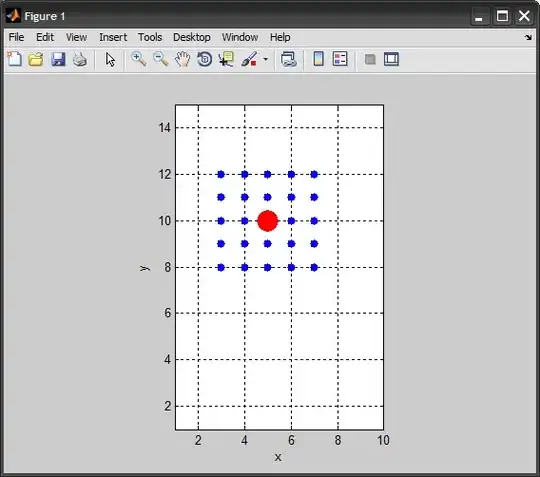
Following the discussion in the comments, I am adding another solution. For a given point, we compute the neighboring points within a specified number of steps (radius if you will). This is shown for both the 2D and the 3D case.
2D matrix
siz = [10 15]; %# matrix size
p = [5 10]; %# 2D point location
%# neighboring points
k = 2; %# radius size
[sx,sy] = ndgrid(-k:k,-k:k); %# steps to get to neighbors
xy = bsxfun(@plus, p, [sx(:) sy(:)]); %# add shift
xy = bsxfun(@min, max(xy,1), siz); %# clamp coordinates within range
xy = unique(xy,'rows'); %# remove duplicates
xy(ismember(xy,p,'rows'),:) = []; %# remove point itself
%# show solution
figure
line(p(1), p(2), 'Color','r', ...
'LineStyle','none', 'Marker','.', 'MarkerSize',50)
line(xy(:,1), xy(:,2), 'Color','b', ...
'LineStyle','none', 'Marker','.', 'MarkerSize',20)
grid on, box on, axis equal
axis([1 siz(1) 1 siz(2)])
xlabel x, ylabel y
3D matrix
siz = [10 15 8]; %# matrix size
p = [5 10 4]; %# 3D point location
%# neighboring points
k = 2; %# radius size
[sx,sy,sz] = ndgrid(-k:k,-k:k,-k:k); %# steps to get to neighbors
xyz = bsxfun(@plus, p, [sx(:) sy(:) sz(:)]); %# add shift
xyz = bsxfun(@min, max(xyz,1), siz); %# clamp coordinates within range
xyz = unique(xyz,'rows'); %# remove duplicates
xyz(ismember(xyz,p,'rows'),:) = []; %# remove point itself
%# show solution
figure
line(p(1), p(2), p(3), 'Color','r', ...
'LineStyle','none', 'Marker','.', 'MarkerSize',50)
line(xyz(:,1), xyz(:,2), xyz(:,3), 'Color','b', ...
'LineStyle','none', 'Marker','.', 'MarkerSize',20)
view(3), grid on, box on, axis equal
axis([1 siz(1) 1 siz(2) 1 siz(3)])
xlabel x, ylabel y, zlabel z
HTH
Amro
- 123,847
- 25
- 243
- 454
-
-
@Amro, thanks for your code Amro! I'm trying to optimize a function that is taking to much time to compute and it's based on circular neighboorhoods. More precisely, I've different circular neighboorhoods and inside of each of them, I want to compute the mean, std, and median of the pixel values underlying them. This is done in the context of the DoG scale representation. The size of the circular filter is given by radius associated at a specific scale (`scalespace_radii`): http://pastebin.com/WkS5ShMf Any suggestion on how to improve the computation is welcomed! – Tin Feb 11 '14 at 22:57
-
@Tin: I took a look but it seems to me that the triple-loops in the code are unavoidable! Trying to vectorize those for-loops will only result in creating huge intermediate matrices (for example by calculating the `distance` for all pixels at once in a vectorized call). I made some slight changes here and there to improve performance (like better preallocation, adding an early-exit test, and moving some computations outside the loops). It will be easier to show my version if you create a new question page first :) – Amro Feb 12 '14 at 00:50
-
@Tin: In the end this sort of calculation is computationally heavy, and the only way I could think of for truly improving performance is to write a MEX-function (implemented in C/C++ language).. – Amro Feb 12 '14 at 00:51