I plot here values over length for a chromosome
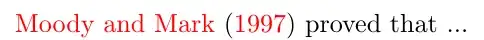
The middle region without points contains no data and should not get a loess line. How can I modify my code to stop the loess line over this region? The data is continuous but I could add lines to mark the blank region with some special value or add a column with a label?? but how to use this in the command?
my current command:
library(IDPmisc)
# plot settings (edit here)
spanv<-0.05
pointcol1="#E69F00"
pointcol2="#56B4E9"
pointcol3="#009E73"
points=20
linecol="green"
xlabs=paste(onechr, " position", " (loess-span=", spanv, ")", sep="")
data1<-NaRV.omit(data[,c(2,7)]) # keep only x and y for the relevant data
# and clean NA and Inf
ylabs='E / A - ratio'
p1<-ggplot(data1, aes(x=start, y=E.R)) +
ylim(0,5) +
geom_point(shape=points, col=pointcol1, na.rm=T) +
geom_hline(aes(yintercept=1, col=linecol)) +
geom_smooth(method="loess", span=spanv, fullrange=F, se=T, na.rm=T) +
xlab(xlabs) +
ylab(ylabs)