You don't need networkX to solve the problem, numpy can do it if you understand the math behind it. A undirected, unweighted graph can always be represented by a [0,1] adjacency matrix. nth powers of this matrix represent the number of steps from (i,j) after n steps. We can work with a Markov matrix, which is a row normalized form of the adj. matrix. Powers of this matrix represent a random walk over the graph. If the graph is small, you can take powers of the matrix and look at the index (start, end) that you are interested in. Make the final state an absorbing one, once the walk hits the spot it can't escape. At each power n you get probability that you'll have diffused from (i,j). The hitting time can be computed from this function (as you know the exact hit time for discrete steps).
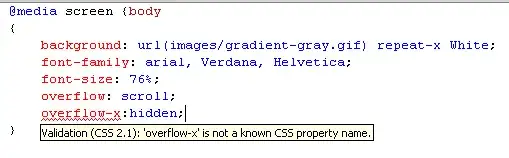
Below is an example with a simple graph defined by the edge list. At the end, I plot this hitting time function. As a reference point, this is the graph used:
from numpy import *
hit_idx = (0,4)
# Define a graph by edge list
edges = [[0,1],[1,2],[2,3],[2,4]]
# Create adj. matrix
A = zeros((5,5))
A[zip(*edges)] = 1
# Undirected condition
A += A.T
# Make the final state an absorbing condition
A[hit_idx[1],:] = 0
A[hit_idx[1],hit_idx[1]] = 1
# Make a proper Markov matrix by row normalizing
A = (A.T/A.sum(axis=1)).T
B = A.copy()
Z = []
for n in xrange(100):
Z.append( B[hit_idx] )
B = dot(B,A)
from pylab import *
plot(Z)
xlabel("steps")
ylabel("hit probability")
show()