Anyone encountered this difficulty with kernlab regression? It seems like it's losing some scaling factors or something, but perhaps I'm calling it wrong.
library(kernlab)
df <- data.frame(x=seq(0,10,length.out=1000))
df$y <- 3*df$x + runif(1000) - 3
plot(df)
res <- ksvm(y ~ x, data=df, kernel='vanilladot')
lines(df$x, predict(res), col='blue', lwd=2)
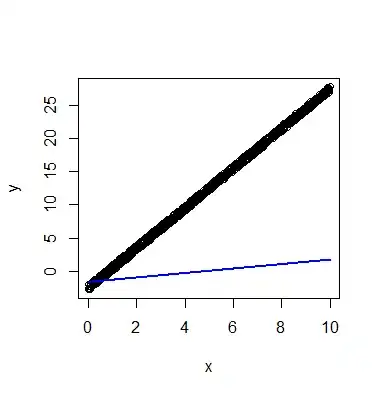
With this toy example I can get reasonable results if I explicitly pass newdata=df, but with my real data I've found no such workaround. Any insight?