Don't forget to set the 'physics engine' of the network graph, and also use the value (not label) parameter when setting the weights of the edges
For example, consider a network visualization showing the frequency of bigrams in a famous piece of English literature, say Shakespeare's "To be or not to be" soliloquy from Hamlet:
from collections import defaultdict
import re
hamlet_speech = # [See link above]
shakespeare_letters = re.sub("[',.;:\-\—?\n ]", "", hamlet_speech.upper())
bigrams = [
shakespeare_letters[i : i + 2]
for i in range(len(shakespeare_letters) - 1)
]
freqs = defaultdict(int)
for xy in bigrams:
freqs[xy] += 1
df = pd.DataFrame(
[[*xy] + [w] for xy, w in freqs.items()], columns=["from", "to", "weight"]
)
df.sort_values(by="weight", inplace=True, ascending=False)
df = df[df.weight > 3]
df
gives:
from to weight
10 T H 53
16 H E 33
35 N D 19
0 T O 18
55 O F 17
.. ... .. ...
226 R D 4
241 L L 4
21 I O 4
8 T T 4
166 P A 4
[99 rows x 3 columns]
Note: I've included only the most frequent (occurrence > 3) pairs of subsequent letters for the sake of simplifying this example.
Unsurprisingly, the most principal results are:
- "TH" (e.g., as in "the"...) is the most common bigram,
- followed by "HE" (also, as in, "the"...).
Let's see how pyvis visually represents this:
import pandas as pd
from IPython.display import display, HTML
from pyvis.network import Network
got_net = Network(
notebook=True,
cdn_resources="remote",
height="500px",
width="100%",
bgcolor="white",
font_color="red",
)
# set the physics layout of the network
got_net.repulsion()
got_data = df
sources = got_data["from"]
targets = got_data["to"]
weights = got_data["weight"]
edge_data = zip(sources, targets, weights)
for e in edge_data:
src = e[0]
dst = e[1]
w = e[2]
got_net.add_node(src, src, title=src)
got_net.add_node(dst, dst, title=dst)
got_net.add_edge(src, dst, value=w)
neighbor_map = got_net.get_adj_list()
# add neighbor data to node hover data
for node in got_net.nodes:
node["title"] += "\nNeighbors:\n"
neighbor_distances = {}
for neighbor in neighbor_map[node["id"]]:
bigram = node["id"] + neighbor
dist = freqs[bigram]
neighbor_distances[neighbor] = dist
for n, d in sorted(
neighbor_distances.items(), key=lambda kv: kv[1], reverse=True
):
node["title"] += f"{n}: {d}\n"
node["value"] = len(neighbor_map[node["id"]])
got_net.show("network.html")
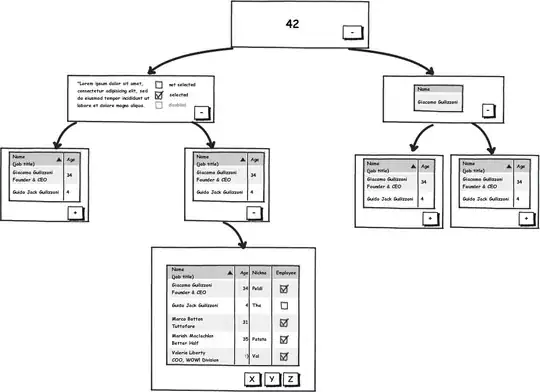
Clearly, a critical consideration with such a type of "network" visualization is the reality that individual nodes can be simultaneously a place of "from" and "to". And thus, when visualizing the "weight" of any connection between two individual nodes, it must be taken into consideration the weight in both directions. This can become obviously quite complex pretty quickly as the number of nodes and possible pairwise paths permutationally increase.
Nonetheless pyvis handles this excellently, by making the network interactive and also by visually representing the strength, if you will, (i.e., the weight) of node interconnections by both their overall placement in the network relative to other nodes but also the width of their edges.
 .
.