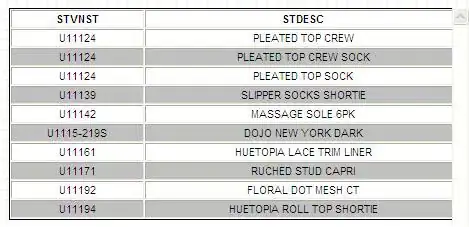
I am trying to join two tables based on a code created within each table that identifies a prescribed drug. The problem is that the drug code sometimes has additional numbers at the end in one table. See ex:
- ncdnum table 1: 4988472401
- ncdnum table 2: 49884724
is there a way to join these two tables so that table 1 is joined by table 2 with that base ndcnum code.
i tried a left join but since none of the values equal each other nothing was joined.
TopNDCCodes <- left_join(ndcnum,
ndccodes,
by = c("ndcnum" = "new_NDC_Product"))