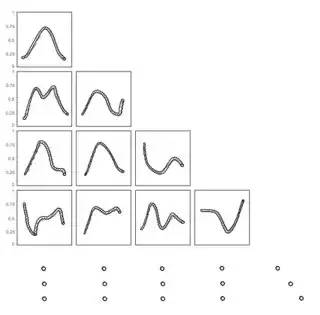
This is a bit different from a standard pairs plot, such as you might produce with GGally::ggpairs, since the x axis is constant across all plots.
If you want to build this yourself then you could automate using the patchwork package.
First, work out the row and column numbers that are going to appear in your plot, and what their names would be if they were columns in your data frame, even for the blank spaces in your plot that do not have a corresponding column in your data frame:
library(ggplot2)
library(patchwork)
n <- which(choose(seq(20), 2) == ncol(df) - 1)
pairs <- apply(expand.grid(seq(n-1) + 1, seq(n - 1)), 1, function(x) {
paste0("r_{", x[2], ",", x[1], "}") })
pairs <- c(t(matrix(pairs, nrow = n - 1)))
pairs
#> [1] "r_{1,2}" "r_{2,2}" "r_{3,2}" "r_{4,2}" "r_{1,3}" "r_{2,3}" "r_{3,3}"
#> [8] "r_{4,3}" "r_{1,4}" "r_{2,4}" "r_{3,4}" "r_{4,4}" "r_{1,5}" "r_{2,5}"
#> [15] "r_{3,5}" "r_{4,5}"
Now we can create a plot for member of pairs. If it does not exist as a column in the data frame, we create an empty plot. If it is on the top row or the rightmost column, we add a single facet. If it is on the bottom or right, we use axis text, and remove it otherwise:
plots <- lapply(pairs, function(i) {
is_bottom <- grepl(paste0(",", n), i)
is_right <- grepl(paste0(n - 1, ","), i)
is_left <- grepl("1,", i)
is_top <- grepl(",2", i)
is_col <- i %in% names(df)
rowname <- sub("^r_\\{(\\d+),.*$", "\\1", i)
colname <- sub("^.*,(\\d+).*$", "\\1", i)
if(!is_col) {
p <- ggplot(df, aes(X, runif(length(x)))) + geom_blank()
} else {
p <- ggplot(df, aes(X, .data[[i]])) +
geom_line(linewidth = 1.5, color = "gray50", lineend = "round")
}
p <- p + scale_y_continuous(NULL, limits = c(0, 1)) +
scale_x_continuous(NULL, 1:10) +
theme_classic()
f <- quote(.~.)
if(is_top) f[3] <- rowname
if(is_right) f[2] <- colname
if(is_top | is_right) p <- p + facet_grid(f)
p + theme(axis.text.x = if(is_bottom) element_text() else element_blank(),
axis.title.x = if(is_bottom) element_text() else element_blank(),
axis.text.y = if(is_left) element_text() else element_blank(),
panel.border = if(is_col) element_rect(fill = NA) else element_blank(),
axis.ticks = if(is_col) element_line() else element_blank(),
axis.line = element_blank())
})
To plot it, we can do:
Reduce(`+`, plots) +
plot_annotation(caption = "X", theme = theme(
plot.caption = element_text(hjust = 0.5, size = 12)))
EDIT
To plot a facet with the column name above each panel, change the creation of plots in the above code to:
plots <- lapply(pairs, function(i) {
is_bottom <- grepl(paste0(",", n), i)
is_right <- grepl(paste0(n - 1, ","), i)
is_left <- grepl("1,", i)
is_top <- grepl(",2", i)
is_col <- i %in% names(df)
if(!is_col) {
p <- ggplot(df, aes(X, runif(length(X)))) + geom_blank()
} else {
p <- ggplot(df, aes(X, .data[[i]])) +
geom_line(linewidth = 1.5, color = "gray50", lineend = "round")
}
p <- p + scale_y_continuous(NULL, limits = c(0, 1)) +
scale_x_continuous(NULL, 1:10) +
theme_classic()
f <- quote(.~.)
f[3] <- i
if(is_col) p <- p + facet_grid(f)
p + theme(axis.text.x = if(is_bottom) element_text() else element_blank(),
axis.title.x = if(is_bottom) element_text() else element_blank(),
axis.text.y = if(is_left) element_text() else element_blank(),
panel.border = if(is_col) element_rect(fill = NA) else element_blank(),
axis.ticks = if(is_col) element_line() else element_blank(),
axis.line = element_blank())
})
And when plotted, this produces
Data used
set.seed(1)
df <- cbind(X = 1:10,
replicate(10, sort(runif(10))) |>
as.data.frame() |>
setNames(combn(1:5, 2, FUN = \(x) paste0("r_{", x[1], ",", x[2], "}"))))
df
#> X r_{1,2} r_{1,3} r_{1,4} r_{1,5} r_{2,3} r_{2,4} r_{2,5}
#> 1 1 0.06178627 0.1765568 0.01339033 0.1079436 0.0233312 0.07067905 0.08424691
#> 2 2 0.20168193 0.2059746 0.12555510 0.1862176 0.4772301 0.09946616 0.25801678
#> 3 3 0.26550866 0.3800352 0.21214252 0.4112744 0.5297196 0.24479728 0.29360337
#> 4 4 0.37212390 0.3841037 0.26722067 0.4820801 0.5530363 0.31627171 0.33239467
#> 5 5 0.57285336 0.4976992 0.34034900 0.4935413 0.6470602 0.40683019 0.45906573
#> 6 6 0.62911404 0.6870228 0.38238796 0.5995658 0.6927316 0.43809711 0.47854525
#> 7 7 0.66079779 0.7176185 0.38611409 0.6684667 0.7323137 0.47761962 0.65087047
#> 8 8 0.89838968 0.7698414 0.65167377 0.7237109 0.7829328 0.51863426 0.76631067
#> 9 9 0.90820779 0.7774452 0.86969085 0.7942399 0.7893562 0.66200508 0.87532133
#> 10 10 0.94467527 0.9919061 0.93470523 0.8273733 0.8209463 0.86120948 0.91287592
#> r_{3,4} r_{3,5} r_{4,5}
#> 1 0.3337749 0.1216919 0.05893438
#> 2 0.3390729 0.1433044 0.23962942
#> 3 0.3466835 0.2026923 0.41008408
#> 4 0.3899895 0.2454885 0.45527445
#> 5 0.4763512 0.3253522 0.60493329
#> 6 0.7773207 0.3999944 0.64228826
#> 7 0.8394404 0.4346595 0.77891468
#> 8 0.8643395 0.7111212 0.79730883
#> 9 0.8921983 0.7125147 0.81087024
#> 10 0.9606180 0.7570871 0.87626921
Created on 2023-07-30 with reprex v2.0.2