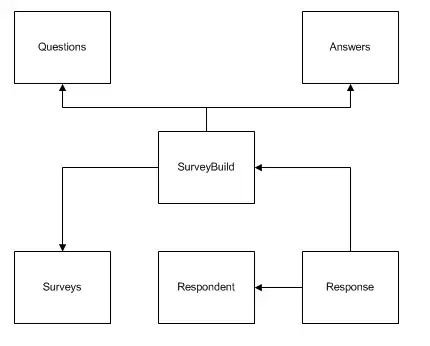
Here is code from an example I was viewing from a mix0mics package about PLS-DA.
library(mixOmics)
data(srbct)
X <- srbct$gene
Y <- srbct$class
final.plsda.srbct <- plsda(X,Y, ncomp = 3)
plotIndiv(final.plsda.srbct, ind.names = FALSE, legend=TRUE,
comp=c(1,2), ellipse = TRUE,
title = 'PLS-DA on SRBCT comp 1-2',
X.label = 'PLS-DA comp 1', Y.label = 'PLS-DA comp 2')
The code generates a figure like this from the PlotIndiv() code line. I want to clean up the graph a bit and remove the gridlines, and only have dashed lines over the X=0 line and Y=0, and to remove the awkward gray box at the top of the figure.
I tried to do title=NULL, but that wouldn't remove that portion. And I though doing something like theme_classic() may change how the figure would look, but still having difficulty to adjust this after looking online sources.
Any feedback on how to change the aesthetics of a plot would be appreciated please.
Thank you.