In R, and RStudio, I have run his code:
#ordinary kriging
mydata.krig <- krige(Clay~1, train_data, newdata=mask, vgm)
names(mydata.krig)
names(mydata.krig)[1] <- "Clay.pred"
names(mydata.krig)
min(mydata.krig$Clay.pred, na.rm=T); max(mydata.krig$Clay.pred, na.rm=T)
ggplot() +
geom_stars(data = mydata.krig["Clay.pred"]) +
scale_fill_gradient(low = "yellow", high = "dark blue", limits = c(15.5486,61.41131)) +
geom_sf(data = thessaly_smus, color = "black", fill = NA, size = 1) +
labs(title = "%Clay Predicted Values - Ordinary Kriging, OK")
With ggplot, I have also acquired a map of the predicted values of %Clay. I want to extract this map as a raster file from R in order to open it in a GIS, but only the area of interest (coloured polygons). Can you provide me accurate code for this?
Updated* With the help of @I_O, I wrote the code as you can see in comments, but again this is not my outcome that I want to take.I took the following outcome: 
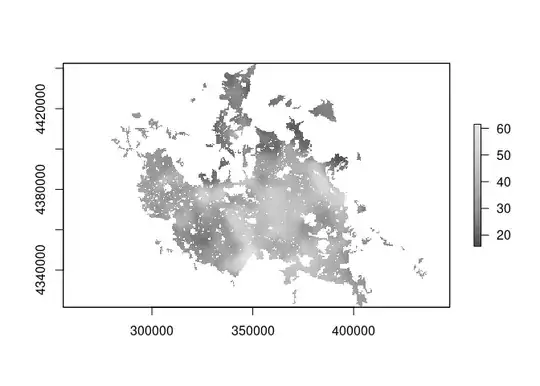
But I want to create a raster file like this that will contain the clay predictions: