I have a plot with the labels on the y axis (after using coord_flip) Here is a reproducible example
Names <- c("ExampleA","ExampleB","ExampleC","ExampleD", "ExampleE", "ExampleF","ExampleG", "ExampleH")
Counts <- c(4,3,2,1,-1,-2,-3,-4)
Type <- c("X","X","X","X", "Y","Y","Y","Y")
df <-data.frame(Names,Counts, Type)
ggplot(df, aes(x=reorder(Names,Counts), y=Counts, fill=Type))+
geom_col()+
coord_flip()+
theme_minimal()
I want the labels to be written next to the bars. Meaning that "ExampleA/B/C/D" etc. should be written right next to the bars and not listed on the y axis. Or in visuals: What I have
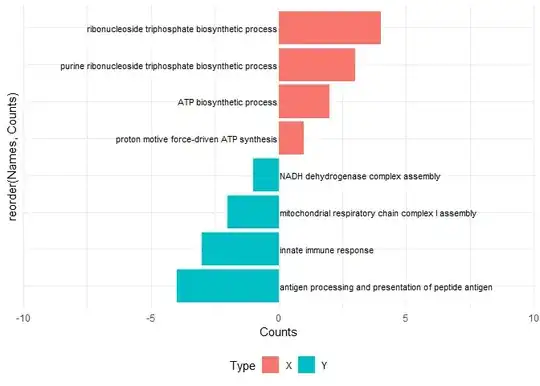
What I want
 (Credit Li et al. 2021 https://doi.org/10.3389/fimmu.2021.694801)
(Credit Li et al. 2021 https://doi.org/10.3389/fimmu.2021.694801)
I tried: ggplot: put axis text inside plot which didn't work.
This
geom_label(label=Names, parse=TRUE)
is close but not really what I want (and also for the real data only works when I type out every single name into a vector which I'd like to avoid if not nescessary). I was hoping that there is something along the lines of
geom_label(label=Names, position="inside")
or
scale_x_discrete(position = "inside")
would be possible, but thus far I could not find anything.