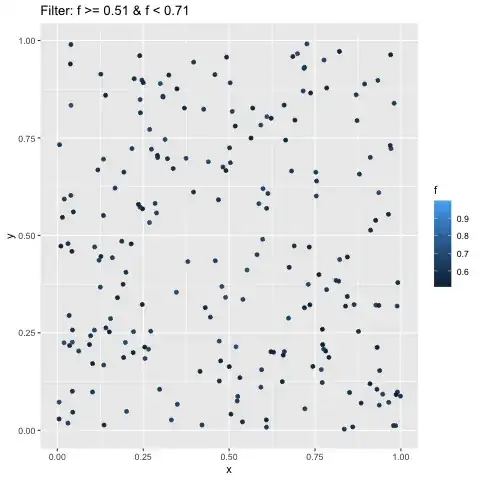
I am trying to summarize the Pearson correlation index between x and y per facet (grouping) z and per frame by transition_filter
data.frame(x = runif(300), y = runif(300), z = runif(300), g = rep(c("a", "b", "c"), each = 100)) %>%
ggplot(aes(x = x, y = y, color = z)) + geom_point() +
facet_wrap(. ~ g) +
transition_filter(transition_length = 1, filter_length = 1,
z >= 0.5 & z < 0.6,
z >= 0.6 & z < 0.7,
z >= 0.7 & z < 0.8,
z >= 0.8 & z < 0.9,
z >= 0.9 & z < 1) +
geom_text(aes(label = paste0("Cor = ", cor(x, y))), x = 0.25, y = 1) +
ggtitle('{closest_expression}')
I can get the correlation indexes displayed though they appear to be the same across facets and frames:
What is the correct way of doing this?