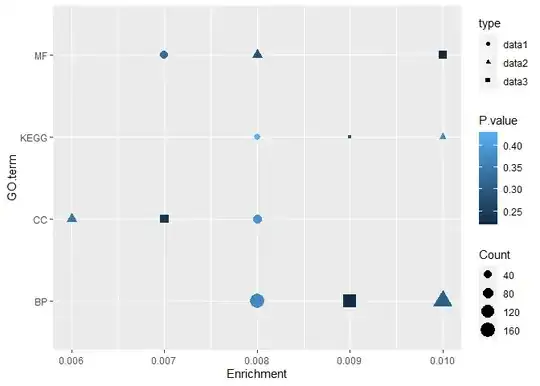
I have three datasets which I would like combine in one scatter plot. The data sets are: data 1 -
| GO term | Count | Enrichment | P value |
|---|---|---|---|
| BP | 163 | 0.008 | 0.37 |
| MF | 48 | 0.007 | 0.33 |
| CC | 58 | 0.008 | 0.39 |
| KEGG | 27 | 0.008 | 0.43 |
data 2 -
| GO term | Count | Enrichment | P value |
|---|---|---|---|
| BP | 167 | 0.01 | 0.31 |
| MF | 50 | 0.008 | 0.29 |
| CC | 50 | 0.006 | 0.34 |
| KEGG | 23 | 0.01 | 0.37 |
data 3 -
| GO term | Count | Enrichment | P value |
|---|---|---|---|
| BP | 123 | 0.009 | 0.22 |
| MF | 44 | 0.01 | 0.22 |
| CC | 50 | 0.007 | 0.24 |
| KEGG | 14 | 0.009 | 0.28 |
## to reproduce
data_1 <- structure(list(GO.term = c("BP", "MF", "CC", "KEGG"), Count = c(163L,
48L, 58L, 27L), Enrichment = c(0.008, 0.007, 0.008, 0.008), P.value = c(0.37,
0.33, 0.39, 0.43)), class = "data.frame", row.names = c(NA, 4L
))
data_2 <- structure(list(GO.term = c("BP", "MF", "CC", "KEGG"), Count = c(167L,
50L, 50L, 23L), Enrichment = c(0.01, 0.008, 0.006, 0.01), P.value = c(0.31,
0.29, 0.34, 0.37)), class = "data.frame", row.names = c(NA, 4L
))
data_3 <- structure(list(GO.term = c("BP", "MF", "CC", "KEGG"), Count = c(123L,
44L, 50L, 14L), Enrichment = c(0.009, 0.01, 0.007, 0.009), P.value = c(0.22,
0.22, 0.24, 0.28)), class = "data.frame", row.names = c(NA, 4L
))
For data 1, I created a scatterplot with ggplot(data, aes(x=Fold.Enrichment, y=GO.Term, color=PValue)) + geom_point(aes(size=Count))
Now I want to combine all the data sets in one plot. Is it possible with scatterplot or do I need to change the graph type?