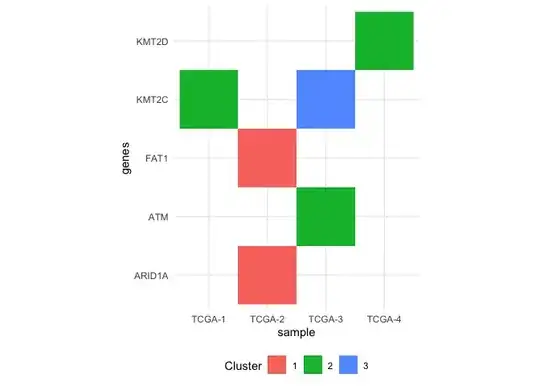
I have a data looks like this.
Genes Sample cluster
1: ARID1A TCGA-2 cluster 1
2: FAT1 TCGA-2 cluster 1
3: KMT2C TCGA-1 cluster 2
4: KMT2C TCGA-3 cluster 3
5: ATM TCGA-3 cluster 2
6: KMT2D TCGA-4 cluster 2
I am wondering is there any way to create a heat map of this kind of data in R. I already tried looking into pheatmap package but couldnot find any solution .