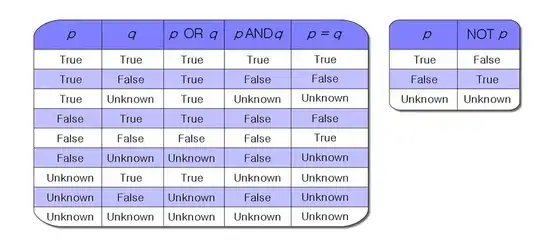
I need to recode an equivalent of the NumPy interp function. In order to do that, I thought of building a pairwise distance matrix, and then using this matrix to define "weights" that I use to multiply the target values.
def interp_diff(x, xp, fp):
dxp = np.diff(xp)
dxp = np.concatenate(([dxp[0]], dxp))
pairwise_distance = np.abs(x[:, np.newaxis] - xp) # Calculate the absolute differences between x and xp
# Add a small epsilon to pairwise distances to avoid division by zero
epsilon = 1e-8
pairwise_distance += epsilon
weights = 1 / pairwise_distance # Calculate the weights based on the differences
# Normalize the weights
weights /= np.sum(weights, axis=1)[:, np.newaxis]
# Apply hardness threshold to the weights
weights = np.where(weights > 0, weights, 0)
return np.dot(weights, fp)
This produces expected results when xp values are placed on a regular grid, but it does not work if xp values are not evenly spaced. How can I make this work for any spacing of xp?
The constraint I have is that I can't use index related methods (argsort, argwhere, searchsorted, etc...). Which is what makes it a bit challenging.
Example usage on a regular grid:
x_np = np.linspace(0, 5, 4)
y_np = np.sin(x_np)
x_i = x_np
x_i = np.linspace(0, 5, 10)
y_i = interp_diff(x_i, xp=x_np, fp=y_np)
ax = plt.figure().gca()
ax.scatter(x_np, y_np)
ax.scatter(x_i, y_i, marker='+', s=30, alpha=0.7)
ax.plot(x_i, y_i)
plt.show()
Produces the expected result:
However, switching to non evenly spaced grid:
def sinspace(start, stop, num):
ones = 0 * start + 1
return start + (stop - start) * (1 - np.cos(np.linspace(
0 * ones,
np.pi / 2 * ones,
num
)))
x_np = sinspace(0, 5, 4)
y_np = np.sin(x_np)
x_i = x_np
x_i = np.linspace(0, 5, 10)
y_i = interp_diff(x_i, xp=x_np, fp=y_np)
ax = plt.figure().gca()
ax.scatter(x_np, y_np)
ax.scatter(x_i, y_i, marker='+', s=30, alpha=0.7)
ax.plot(x_i, y_i)
plt.show()
This is what I get: