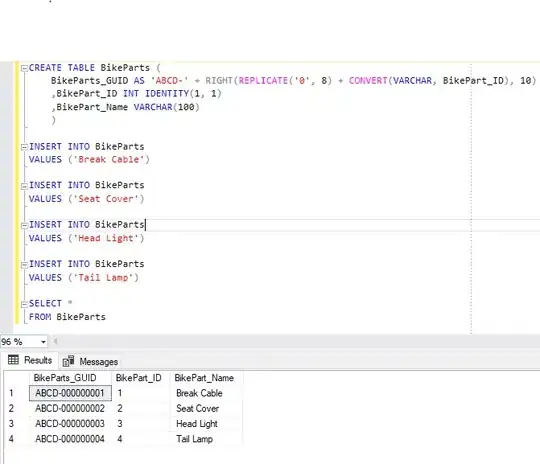
I have a network, and a focal node within the network. I have calculated the shortest paths to all other nodes from that node, and I'm trying to figure out how to make a sub-graph of the original network from that shortest path calculation.
The goal is to be able to show the all the paths from my focal node to any other node that match a criteria (e.g. that the path is less than 50m or longer than 100m but shorter than 500m).
library(sfnetworks)
library(sf)
library(tidygraph)
library(dplyr)
library(ggplot2)
# the base network
net <- as_sfnetwork(roxel, directed = FALSE) %>%
st_transform(3035) %>%
activate("edges") %>%
mutate(weight = edge_length())
# the network with the focal point
ggplot() +
geom_sf(data = net %>%
activate("edges") %>%
st_as_sf(), colour = "grey90") +
geom_sf(data = net %>%
activate("nodes") %>%
st_as_sf()) +
geom_sf(data = net %>%
activate("nodes") %>%
slice(495) %>%
st_as_sf(), size = 3.5, fill = "orange",
colour = "black", shape = 21) +
theme_void()
I can get the shortest paths using the sfnetworks wrapper or directly from igraph:
short_paths_sf <- st_network_paths(net %>% activate(nodes),
from = 495, to = c(1:701), weights = "weight")
short_paths_ig <- igraph::shortest_paths(
graph = net,
from = 495,
to = c(1:701),
output = "both",
weights = net %>% activate(edges) %>% pull(weight)
)
But I think I'm missing two key parts here. 1) Making the subgraph from the net object and containing just the shortest paths and 2) then transforming the subgraph so I can do operations on the length of the whole path not just the weight of an edge.
I know I can get the length of a single path between two points with something like
single_path <- st_network_paths(net %>% activate(nodes),
from = 495,
to = 701,
weights = "weight") %>%
pull(edge_paths) %>%
unlist()
net %>%
activate(edges) %>%
slice(single_path) %>%
st_as_sf() %>%
st_combine() %>%
st_length()
> 482.0495 [m]
Which I can plot with
ggplot() +
geom_sf(data = net %>%
activate("edges") %>%
st_as_sf(), colour = "grey90") +
geom_sf(data = net %>%
activate("nodes") %>%
st_as_sf()) +
geom_sf(data = net %>%
activate("nodes") %>%
slice(495) %>%
st_as_sf(), size = 3.5, fill = "orange",
colour = "black", shape = 21) +
geom_sf(data = net %>%
activate(edges) %>%
slice(single_path) %>%
st_as_sf(), colour = "firebrick") +
theme_void()
But I'm struggling to figure out how to do that for the single-to-many case, and then especially, how to filter those paths such that, for example, I could look at only paths from point 495 that are shorter than 500m.
I tried to apply what the authors here suggest in the shortest paths section:
sub_graph <- net %>%
igraph::subgraph.edges(eids = short_paths_ig$epath %>% unlist()) %>%
as_tbl_graph()
But then it's unclear to me how this will show me my shortest paths?
Any help much appreciated!!