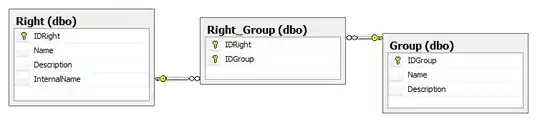
I have created two separate ridgeplots in R using the ggridge package and the stat_density_ridges() function. My dataset includes a series of Latitude and Longitude locations. I have produced one ridgeplot for Latitute and one for Longitude and they are perfectly fine on their own. I have then used the function plot_grid() from the package cowplot to arrange the two plots horizontally. The two plots are aligned in terms of plot area but the x axes are not. Also, the spacing between plots is quite large and I would prefer reducing it rather than editing it manually.
My code is:
#load the necessary libraries
library(readr)
library(ggplot2)
library(ggridges)
library(cowplot)
library(RColorBrewer)
#load the data into a data.frame object
data <- as.data.frame(read_csv(file.choose()))
#View(data)
#calculate min and max latitude and longitude
minlat <- min(data$lat)
maxlat = max(data$lat)
minlon <- min(data$lon)
maxlon = max(data$lon)
#define the color palette to match the number of groups in the dataset
nb.cols <- 18
mycolors <- colorRampPalette(brewer.pal(9, "YlGnBu"))(nb.cols)
#make the latitude ridgeplot
r.plot.lat <-
ggplot(data, aes(x = lat, y = species_group, fill = species_group)) +
stat_density_ridges(
alpha = 0.75,
scale = 5,
panel_scaling = T,
size = 0.6,
linetype = 1,
colour = "darkgrey",
quantile_lines = F,
rel_min_height = 0.01
) +
theme_ridges(font_size = 18) +
theme(legend.position = "none") +
xlab("Latitude") +
ylab("") +
scale_fill_manual(values = mycolors) +
scale_x_discrete(limits=seq(round(minlat,0),round(maxlat,0), 2))
r.plot.lat
#make the longitude ridgeplot
r.plot.lon <-
ggplot(data, aes(x = lon, y = species_group, fill = species_group)) +
stat_density_ridges(
alpha = 0.75,
scale = 5,
panel_scaling = T,
size = 0.6,
linetype = 1,
colour = "darkgrey",
quantile_lines = F,
rel_min_height = 0.01
) +
theme_ridges(font_size = 18) +
theme(legend.position = "none") +
xlab("Longitude") +
ylab("")+
theme(axis.text.y=element_blank()) +
scale_fill_manual(values = mycolors) +
scale_x_discrete(limits=seq(round(minlon,1),round(maxlon,1), 5))
r.plot.lon
#arrange the plots horizontally
plot_grid(r.plot.lat, NULL, r.plot.lon, align = "hv", labels=c("AUTO"), rel_widths = c(3, 0, 3), nrow=1)
The resulting plot is (I have blurred the Y axes on purpose, the plot is going into a paper to be submitted for publication):
I thought it was a problem of bandwidth so I have specified the same value for each plot in the bandwidth argument in the stat_density_ridges() function. Still not aligned.
EDIT Following @Quinten request, here is an example of 60 random records from my dataset.
structure(list(species_group = c("Species_1", "Species_2", "Species_3",
"Species_4", "Species_5", "Species_2", "Species_6", "Species_7",
"Species_2", "Species_2", "Species_8", "Species_9", "Species_8",
"Species_9", "Species_8", "Species_10", "Species_2", "Species_6",
"Species_3", "Species_1", "Species_1", "Species_1", "Species_10",
"Species_8", "Species_2", "Species_2", "Species_11", "Species_2",
"Species_2", "Species_3", "Species_3", "Species_2", "Species_2",
"Species_1", "Species_5", "Species_6", "Species_3", "Species_11",
"Species_5", "Species_8", "Species_2", "Species_2", "Species_8",
"Species_2", "Species_11", "Species_12", "Species_9", "Species_3",
"Species_5", "Species_13", "Species_11", "Species_9", "Species_3",
"Species_2", "Species_1", "Species_5", "Species_1", "Species_3",
"Species_7", "Species_3"), lat = c(39.29652333333, 38.69591833333,
43.78736, 42.88574333333, 38.51973, 40.06572, 37.36825, 41.06519666667,
38.18816166667, 36.31976333333, 42.37949166667, 38.66838666667,
40.72673833333, 36.75656833333, 40.47298333333, 41.720205, 36.14439333333,
36.64385666667, 35.40855333333, 40.5198, 42.94889333333, 35.99670666667,
39.967185, 39.37684333333, 39.68108666667, 35.84022166667, 39.45906833333,
38.6217, 36.79680166667, 39.819615, 36.36867666667, 40.399535,
36.28853, 42.11886, 40.43070833333, 39.200125, 36.90197, 37.47650833333,
38.10325666667, 41.64378333333, 34.68400166667, 40.42848333333,
38.52368166667, 36.15304166667, 36.88089333333, 38.7511, 37.15612666667,
41.83358833333, 39.90101833333, 39.90281, 36.207415, 44.05422166667,
44.02940666667, 36.873425, 39.91606166667, 42.93375166667, 39.68787833333,
38.57978166667, 39.72445333333, 38.77966), lon = c(5.92169333333,
10.60350166667, 9.29786666667, 7.91347833333, 17.63931166667,
3.12386166667, 23.40353, 12.51722833333, 11.13067333333, -6.91365,
15.645445, 10.63217166667, 1.70525666667, -1.71539666667, 1.47485833333,
3.81735666667, -2.61197833333, -7.662125, 12.72044666667, 5.24996,
10.23093833333, -5.369945, 17.42619666667, 4.91467166667, 1.518235,
-6.721575, 0.70451333333, 19.47572333333, -1.51764666667, 17.444935,
-6.83558166667, 6.89818, -6.96372, 6.03615833333, 3.20266, 0.79276166667,
-0.75958833333, 2.97557333333, 1.92862666667, 19.300375, 29.77435166667,
6.52557, 19.43436333333, -4.59611666667, 0.77969, 2.05171833333,
-0.81246833333, 7.34824, 1.64478333333, 11.36721, -3.84277166667,
8.53026833333, 9.25958, 0.78068, 1.62469, 3.64922666667, 0.81624166667,
17.63248166667, 10.09109666667, 10.969315), type = c("groups",
"groups", "species", "species", "groups", "groups", "species",
"groups", "groups", "groups", "groups", "groups", "groups", "groups",
"groups", "groups", "groups", "species", "species", "groups",
"groups", "groups", "groups", "groups", "groups", "groups", "groups",
"groups", "groups", "species", "species", "groups", "groups",
"groups", "groups", "species", "species", "groups", "groups",
"groups", "groups", "groups", "groups", "groups", "groups", "groups",
"groups", "species", "groups", "species", "groups", "groups",
"species", "groups", "groups", "groups", "groups", "species",
"groups", "species")), row.names = c(NA, -60L), class = "data.frame")