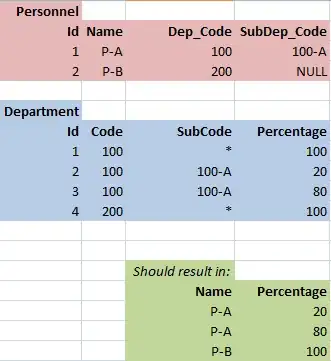
I am making a graph for my data and am using the geom_smooth function in ggplot to make them smooth looking line graphs. I would like to make the graphs colorblind friendly, so would like to make the lines either different line types or the points different shapes. The graphs do not like the code that I am using and displays this graph:
Here is the code that I am using:
ggplot(leach.conc, aes(x = days, y = cl_conc, linetype = treatment, color = factor(treatment, level = c('Liquid', 'Solid', 'KCl', 'Control')))) +
labs(x = "Days since application", y = "Chloride concentration (mg/L)", color = "Treatment") +
geom_smooth() + facet_wrap(.~soil_type) +
scale_x_continuous(breaks = c(0,4,11,18))
Here is a dput for reproducibility
leach.conc <- data.frame(
core = c(
"MS", "MS", "MS", "MS", "ML", "ML", "ML", "ML", "MK", "MK",
"MK", "MK", "MC", "MC", "MC", "MC", "FS", "FS", "FS", "FS", "FL",
"FL", "FL", "FL", "FK", "FK", "FK", "FK", "FC", "FC", "FC", "FC",
"MS", "MS", "MS", "MS", "ML", "ML", "ML", "ML", "MK", "MK", "MK",
"MK", "MC", "MC", "MC", "MC", "FS", "FS", "FS", "FS", "FL", "FL",
"FL", "FL", "FK", "FK", "FK", "FK", "FC", "FC", "FC", "FC", "MS",
"MS", "MS", "MS", "ML", "ML", "ML", "ML", "MK", "MK", "MK", "MK",
"MC", "MC", "MC", "MC", "FS", "FS", "FS", "FS", "FL", "FL", "FL",
"FL", "FK", "FK", "FK", "FK", "FC", "FC", "FC", "FC", "CS", "CL",
"CK", "CC", "PS", "PL", "PK", "PC", "CS", "CL", "CK", "CC", "PS",
"PL", "PK", "PC", "CS", "CL", "CK", "CC", "PS", "PL", "PK", "PC",
"CS", "CL", "CK", "CC", "PS", "PL", "PK", "PC", "CS", "CL", "CK",
"CC", "PS", "PL", "PK", "PC", "CS", "CL", "CK", "CC", "PS", "PL",
"PK", "PC", "CS", "CL", "CK", "CC", "PS", "PL", "PK", "PC", "CS",
"CL", "CK", "CC", "PS", "PL", "PK", "PC", "CS", "CL", "CK", "CC",
"PS", "PL", "PK", "PC", "CS", "CK", "CL", "CC", "PS", "PL", "PK",
"PC", "CS", "CL", "CK", "CC", "PS", "PL", "PK", "PC", "CS", "CL",
"CK", "CC", "PS", "PL", "PK", "PC"
),
core_id = c(
"MS1", "MS1", "MS1", "MS1", "ML1", "ML1", "ML1", "ML1", "MK1",
"MK1", "MK1", "MK1", "MC1", "MC1", "MC1", "MC1", "FS1", "FS1",
"FS1", "FS1", "FL1", "FL1", "FL1", "FL1", "FK1", "FK1", "FK1",
"FK1", "FC1", "FC1", "FC1", "FC1", "MS2", "MS2", "MS2", "MS2",
"ML2", "ML2", "ML2", "ML2", "MK2", "MK2", "MK2", "MK2", "MC2",
"MC2", "MC2", "MC2", "FS2", "FS2", "FS2", "FS2", "FL2", "FL2",
"FL2", "FL2", "FK2", "FK2", "FK2", "FK2", "FC2", "FC2", "FC2",
"FC2", "MS3", "MS3", "MS3", "MS3", "ML3", "ML3", "ML3", "ML3",
"MK3", "MK3", "MK3", "MK3", "MC3", "MC3", "MC3", "MC3", "FS3",
"FS3", "FS3", "FS3", "FL3", "FL3", "FL3", "FL3", "FK3", "FK3",
"FK3", "FK3", "FC3", "FC3", "FC3", "FC3", "CS1", "CL1", "CK1",
"CC1", "PS1", "PL1", "PK1", "PC1", "CS2", "CL2", "CK2", "CC2",
"PS2", "PL2", "PK2", "PC2", "CS3", "CL3", "CK3", "CC3", "PS3",
"PL3", "PK3", "PC3", "CS1", "CL1", "CK1", "CC1", "PS1", "PL1",
"PK1", "PC1", "CS2", "CL2", "CK2", "CC2", "PS2", "PL2", "PK2",
"PC2", "CS3", "CL3", "CK3", "CC3", "PS3", "PL3", "PK3", "PC3",
"CS1", "CL1", "CK1", "CC1", "PS1", "PL1", "PK1", "PC1", "CS2",
"CL2", "CK2", "CC2", "PS2", "PL2", "PK2", "PC2", "CS3", "CL3",
"CK3", "CC3", "PS3", "PL3", "PK3", "PC3", "CS1", "CK1", "CL1",
"CC1", "PS1", "PL1", "PK1", "PC1", "CS2", "CL2", "CK2", "CC2",
"PS2", "PL2", "PK2", "PC2", "CS3", "CL3", "CK3", "CC3", "PS3",
"PL3", "PK3", "PC3"
),
soil_type = rep(
c(
"WSL", "NCL ", "WSL", "NCL ", "WSL", "NCL ", "ESL", "FAH",
"ESL", "FAH", "ESL", "FAH", "ESL", "FAH", "ESL", "FAH", "ESL",
"FAH", "ESL", "FAH", "ESL", "FAH", "ESL", "FAH", "ESL", "FAH",
"ESL", "FAH", "ESL", "FAH"
),
rep(c(16L, 4L), c(6L, 24L))
),
treatment = c(
"Tl", "Tl", "Tl", "Tl", "SM", "SM", "SM", "SM", "KCl", "KCl",
"KCl", "KCl", "Control", "Control", "Control", "Control", "Tl",
"Tl", "Tl", "Tl", "SM", "SM", "SM", "SM", "KCl", "KCl", "KCl",
"KCl", "Control", "Control", "Control", "Control", "Tl", "Tl",
"Tl", "Tl", "SM", "SM", "SM", "SM", "KCl", "KCl", "KCl", "KCl",
"Control", "Control", "Control", "Control", "Tl", "Tl", "Tl",
"Tl", "SM", "SM", "SM", "SM", "KCl", "KCl", "KCl", "KCl", "Control",
"Control", "Control", "Control", "Tl", "Tl", "Tl", "Tl", "SM",
"SM", "SM", "SM", "KCl", "KCl", "KCl", "KCl", "Control", "Control",
"Control", "Control", "Tl", "Tl", "Tl", "Tl", "SM", "SM", "SM",
"SM", "KCl", "KCl", "KCl", "KCl", "Control", "Control", "Control",
"Control", "Tl", "SM", "KCl", "Control", "Tl", "SM", "KCl", "Control",
"Tl", "SM", "KCl", "Control", "Tl", "SM", "KCl", "Control", "Tl",
"SM", "KCl", "Control", "Tl", "SM", "KCl", "Control", "Tl", "SM",
"KCl", "Control", "Tl", "SM", "KCl", "Control", "Tl", "SM", "KCl",
"Control", "Tl", "SM", "KCl", "Control", "Tl", "SM", "KCl", "Control",
"Tl", "SM", "KCl", "Control", "Tl", "SM", "KCl", "Control", "Tl",
"SM", "KCl", "Control", "Tl", "SM", "KCl", "Control", "Tl", "SM",
"KCl", "Control", "Tl", "SM", "KCl", "Control", "Tl", "SM", "KCl",
"Control", "Tl", "KCl", "SM", "Control", "Tl", "SM", "KCl", "Control",
"Tl", "SM", "KCl", "Control", "Tl", "SM", "KCl", "Control", "Tl",
"SM", "KCl", "Control", "Tl", "SM", "KCl", "Control"
),
days = c(
0L, 4L, 11L, 18L, 0L, 4L, 11L, 18L, 0L, 4L, 11L, 18L, 0L, 4L,
11L, 18L, 0L, 4L, 11L, 18L, 0L, 4L, 11L, 18L, 0L, 4L, 11L, 18L,
0L, 4L, 11L, 18L, 0L, 4L, 11L, 18L, 0L, 4L, 11L, 18L, 0L, 4L,
11L, 18L, 0L, 4L, 11L, 18L, 0L, 4L, 11L, 18L, 0L, 4L, 11L, 18L,
0L, 4L, 11L, 18L, 0L, 4L, 11L, 18L, 0L, 4L, 11L, 18L, 0L, 4L,
11L, 18L, 0L, 4L, 11L, 18L, 0L, 4L, 11L, 18L, 0L, 4L, 11L, 18L,
0L, 4L, 11L, 18L, 0L, 4L, 11L, 18L, 0L, 4L, 11L, 18L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L,
4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 11L,
11L, 11L, 11L, 11L, 11L, 11L, 11L, 11L, 11L, 11L, 11L, 11L, 11L,
11L, 11L, 11L, 11L, 11L, 11L, 11L, 11L, 11L, 11L, 18L, 18L, 18L,
18L, 18L, 18L, 18L, 18L, 18L, 18L, 18L, 18L, 18L, 18L, 18L, 18L,
18L, 18L, 18L, 18L, 18L, 18L, 18L, 18L
),
cl_conc = c(
3.6, 18.1, 18.1, 17.4, 3.8, 77.1, 81.4, 66.8, 4.6, 19.4, 22.3, 36.9, 4.1, 1.9,
1.2, 0.6, 18.2, 27.8, 28.3, 28.3, 15, 107.8, 150.3, 94.6, 12.3, 84.8, 53.4,
51.9, 17.3, 9.1, 4.25, 1.9, 3.4, 19.8, 20.7, 20.5, 2.3, 102, 56.7, 47.4, 2.6,
33.4, 15.3, 19.9, 2.9, 2, 1.2, 0.8, 8.1, 37.1, 39.8, 34.8, 4.9, 81.9, 67.5,
56, 5.9, 41.1, 38.3, 30.9, 17.5, 12.4, 6, 3.1, 2.4, 27.8, 27.8, 24.9, 2.6,
79.7, 65.5, 55.2, 2.1, 13.5, 20.4, 24.7, 2, 1.6, 1.2, 0.7, 8.4, 42.7, 40.5,
30.1, 10.3, 121.2, 73.6, 38, 9.4, 53, 38.5, 22.3, 6.6, 4.7, 1.9, 0.85, 4.3,
5.5, 5.6, 7.3, 3.7, 0.9, 2.8, 7.633503186, 5.245095388, 4.9, 6.3, 5.6, 1.4, 5,
5.6, 1.4, 5.7, 15.80504568, 7.6, 6.5, 2.5, 2, 2.6, 1.6, 46.5, 52.6, 32.9, 2.8,
45.1, 1.3, 51.2, 2.6, 47.59251129, 68.3, 38.8, 5.4, 34.1, 66.7, 23.51266468,
0.6, 34.2, 55.7, 23.8, 5, 42.1, 47.9, 44.3, 0.8, 56.23151874, 81.2, 36.1, 1.6,
36.3, 48.2, 35.6, 1.5, 44.8, 80.9, 34.66600908, 3.1, 33.3, 81.5, 20.2, 0.4,
40.1, 66.8, 24.5, 3.6, 39, 68.2, 36, 0.303367677, 31.1, 23.2, 75.7, 0.6, 26.2,
45.3, 21.3, 0.6, 33.76030379, 47.5, 20.5, 1.1, 28.6, 65.9, 18.9, 0.2, 30.2,
65.5, 23.3, 2.7, 23.9, 64, 24.7, 0.1
),
cl_load = c(
0.0072, 0.058825, 0.0543, 0.0609, 0.0076, 0.26985, 0.26455, 0.2171, 0.0092,
0.0582, 0.0669, 0.119925, 0.0082, 0.0057, 0.0036, 0.00195, 0.0364, 0.09035,
0.0849, 0.0849, 0.03, 0.3773, 0.4509, 0.3311, 0.0246, 0.2756, 0.1602, 0.1557,
0.0346, 0.03185, 0.010625, 0.006175, 0.0034, 0.06435, 0.07245, 0.07175,
0.00345, 0.3315, 0.19845, 0.1659, 0.0039, 0.10855, 0.05355, 0.064675,
0.003625, 0.0065, 0.0039, 0.0026, 0.01215, 0.1113, 0.1393, 0.1044, 0.00735,
0.266175, 0.185625, 0.168, 0.00885, 0.113025, 0.105325, 0.0927, 0.02625,
0.0372, 0.018, 0.010075, 0.006, 0.09035, 0.09035, 0.0747, 0.0065, 0.2391,
0.22925, 0.1656, 0.004725, 0.0405, 0.0663, 0.06175, 0.0055, 0.0048, 0.0036,
0.0021, 0.0252, 0.1281, 0.131625, 0.097825, 0.0309, 0.3939, 0.2392, 0.114,
0.0235, 0.17225, 0.125125, 0.0669, 0.0165, 0.015275, 0.006175, 0.0023375,
0.0043, 0.00825, 0.0042, 0.0073, 0.0037, 0.001575, 0.0014, 0.011450255,
0.005245095, 0.0049, 0.007875, 0.0042, 0.00175, 0.005, 0.0112, 0.0014,
0.00285, 0.015805046, 0.0114, 0.008125, 0.00375, 0.0025, 0.00325, 0.0032,
0.11625, 0.14465, 0.0987, 0.0084, 0.0902, 0.0039, 0.1152, 0.00715,
0.118981278, 0.2049, 0.1164, 0.0135, 0.093775, 0.2001, 0.064659828, 0.0018,
0.0855, 0.1671, 0.06545, 0.0125, 0.094725, 0.131725, 0.099675, 0.0024,
0.177129284, 0.2436, 0.1083, 0.0048, 0.1089, 0.15665, 0.1068, 0.0045, 0.1344,
0.2427, 0.11266453, 0.010075, 0.0999, 0.2445, 0.0606, 0.0012, 0.1203, 0.2004,
0.0735, 0.0126, 0.117, 0.2046, 0.117, 0.000985945, 0.0933, 0.0696, 0.2268,
0.0018, 0.0917, 0.15855, 0.0639, 0.00195, 0.101280911, 0.1425, 0.0615,
0.003025, 0.0858, 0.214175, 0.0567, 0.00065, 0.1057, 0.212875, 0.075725,
0.0081, 0.0717, 0.192, 0.0741, 3e-04
)
)