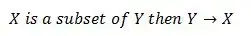I wanna plot my data in an interval between 18:00 pm and 7:00 am ( date as x axis) during a week.
I wanna plot my data along the intervals 18:00 pm and 7:00 am. I've got data gathered during a week every 5 minutes and I've tried to filter out the desired interval but I can't come out with the expected outcome. This is what I've done so far: (I'm new to R by the way)
class(Marzo$date)
Marzo$hms <- format(as.POSIXct(Marzo$date), "%H:%M:%S")
data1 <- Marzo %>% filter(hms >="18:00:00" & hms <="7:00:00") %>% print(n=250)
ggplot() +
geom_line(data= data1, aes(x= date, y= RH3000, colour= "RH3000"),
lwd=1) +
geom_line(data= data1, aes(x= date, y= RH4000, colour= "RH4000"),
lwd=1) +
scale_colour_manual("", breaks=c("RH3000","RH4000"),
values=c("blue4","turquoise1")) +
scale_x_datetime(labels= date_format("%m-%d %H:%M"),
date_breaks= "12 hours")
When I apply that filter this is what I get:
2023-03-13 23:45:00
2023-03-13 23:50:01
2023-03-13 23:55:00
2023-03-14 18:00:00
2023-03-14 18:05:01
Once it gets to 00:00 pm it starts again with the hour 18:00 pm of the next day instead of carrying out till getting till 7:00 am of the next day. Sorry if I was a bit misleading buy Im not English. Any kind of help would come in handy.
Also it would be great to get rid of the straight lines that show up between the non-selected intervals. Graphic: