This required some digging, but I believe I have a working solution.
When you create an instance of the Stargazer class such as your object stargazer, most of the regression results are extracted from the est object which is of type ResultsWrapper (from statsmodels).
Three instance methods called extract_data, _extract_feature, and extract_model_data are called and extract_model_data does a lot of the heavy lifting: it specifically extracts features stored in statsmodels_map, which looks like the following:
statsmodels_map = {'p_values' : 'pvalues', ## ⭠ and this too
'cov_values' : 'params',
'cov_std_err' : 'bse', ## ⭠ we want to modify this
'r2' : 'rsquared',
'r2_adj' : 'rsquared_adj',
'f_p_value' : 'f_pvalue',
'degree_freedom' : 'df_model',
'degree_freedom_resid' : 'df_resid',
'nobs' : 'nobs',
'f_statistic' : 'fvalue'
}
What we can do is create a child class called SuperStargazer that inherits all of the instance methods from Stargazer, and then override the extract_model_data to set cov_std_err using your custom_standard_errors function. Although it's not strictly necessary, we'll set custom_standard_errors as an instance attribute of the SuperStargazer class, as this allows you to use different custom functions if you define different instances of the SuperStargazer class.
Update: we'll also update the p-values as they are related to the new custom standard errors. This involves recalculating the t-values, and then applying the formula: p_value = 2*(1 - t.cdf(abs(t_value), dof))
from statsmodels.base.wrapper import ResultsWrapper
from statsmodels.regression.linear_model import RegressionResults
from scipy.stats import t
from math import sqrt
from collections import defaultdict
from enum import Enum
import numbers
import pandas as pd
class SuperStargazer(Stargazer):
def __init__(self, models, custom_standard_errors, **kwargs):
self.custom_standard_errors = custom_standard_errors
super().__init__(models=models, **kwargs)
def extract_model_data(self, model):
# For features that are simple attributes of "model", establish the
# mapping with internal name (TODO: adopt same names?):
statsmodels_map = {# 'p_values' : 'pvalues',
'cov_values' : 'params',
# 'cov_std_err' : 'bse',
'r2' : 'rsquared',
'r2_adj' : 'rsquared_adj',
'f_p_value' : 'f_pvalue',
'degree_freedom' : 'df_model',
'degree_freedom_resid' : 'df_resid',
'nobs' : 'nobs',
'f_statistic' : 'fvalue'
}
data = {}
for key, val in statsmodels_map.items():
data[key] = self._extract_feature(model, val)
if isinstance(model, ResultsWrapper):
data['cov_names'] = model.params.index.values
endog, exog = model.model.data.orig_endog, model.model.data.orig_exog
custom_std_err_data = self.custom_standard_errors(endog, exog)
data['cov_std_err'] = pd.Series(
index=exog.columns,
data=custom_std_err_data
)
data['t_values'] = data['cov_values'] / data['cov_std_err']
dof = len(endog) - 2
data['p_values'] = pd.Series(
index=data['t_values'].index,
data=2 * (1 - t.cdf(abs(data['t_values']), dof))
)
else:
# Simple RegressionResults, for instance as a result of
# get_robustcov_results():
data['cov_names'] = model.model.data.orig_exog.columns
# These are simple arrays, not Series:
for what in 'cov_values', 'cov_std_err':
data[what] = pd.Series(data[what],
index=data['cov_names'])
data['conf_int_low_values'] = model.conf_int()[0]
data['conf_int_high_values'] = model.conf_int()[1]
data['resid_std_err'] = (sqrt(sum(model.resid**2) / model.df_resid)
if hasattr(model, 'resid') else None)
# Workaround for
# https://github.com/statsmodels/statsmodels/issues/6778:
if 'f_statistic' in data:
data['f_statistic'] = (lambda x : x[0, 0] if getattr(x, 'ndim', 0)
else x)(data['f_statistic'])
return data
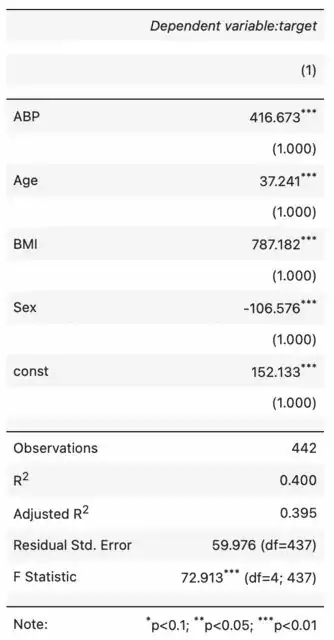
Then when we can create an instance of SuperStargazer, it will render the following table (in JupyterLab in my case, but you can also call stargazer.render_html() to store the html string for later use)
stargazer = SuperStargazer(
models=[est],
custom_standard_errors=custom_standard_errors
)