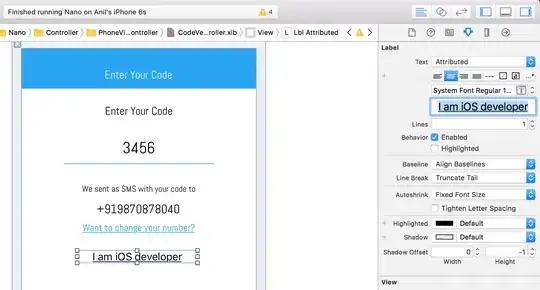
I want to find missing values in a dataframe with both categorical and numerical data. As for the categorical data, one column takes into account the order (here column named "category_with_order"), and the other don't (here column named "category_without_order"). "sugar" and "salt" columns take numerical data as input.
Finally I want to de-encode the whole to retrieve my original headers and the new dataframe imputed with KNN imputer.
What I did is I "One Hot encoded" the categories, but with no distinction between the two categorical columns and I now have two "Nan" columns which is weird :
and then I did a concatenation of my initial dataframe with the categorical variables encoded:
Here is the code I have been using to output these dataframes:
import pandas as pd
import numpy as np
from sklearn.preprocessing import OneHotEncoder
from sklearn.model_selection import train_test_split
from sklearn.impute import KNNImputer
data = {
'category_with_order': ['a', 'b', 'c','d',np.nan],
'category_without_order': ['plant',np.nan,'salad','meat', 'drinks'],
'sugar': ['1',np.nan, '2', '2',np.nan],
'salt': ['1',np.nan, '2', '1',np.nan]
}
df = pd.DataFrame(data1)
ohe = OneHotEncoder()
feature_array = ohe.fit_transform(df[["category_with_order","category_without_order"]]).toarray()
features_labels = ohe.categories_
feature_labels = np.hstack([i.ravel() for i in features_labels])
features = pd.DataFrame(feature_array, columns = feature_labels)
knn = KNNImputer(n_neighbors=1, add_indicator = True)
df_new = pd.concat([df1.reset_index(drop=True), features.reset_index(drop=True)], axis=1)