I am looking at analysis of multiple sets of scanning electron microscope (SEM) images of backscattered electron (BSE) and cathodoluminescence (CL) scans of rock samples.
There are multiple datasets, each of a few hundred images each from a run across a rock sample. Due to the nature of the scanning, there is an offset (which should be approximately constant) across each dataset of <~10%. The alignment is therefore anticipated to be a simple translation of each image.
The nature of the two types of images is that they detect different properties of the rocks, however both detect the edges of the rock grains to differing degrees (very dark areas on BSE images, gray areas with faint outlines on CL images).
I am trying to automate the alignment process prior to performing some segmentation analysis which requires combining results from both images.
Some images have features which I have managed to align using OpenCV findTransformECC, mostly where there is some bright feature in both images, such as this pair (BSE top, CL bottom) which I assume is succeeding based on matching the feature in top right:

When run through the function below, these images align nicely as follows:
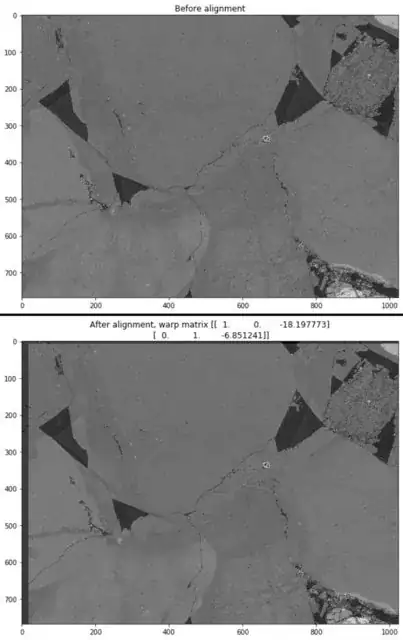
However, most of the images do not have such obvious features and if this was being done manually, it would be done by aligning obvious black areas on the BSE images with faint areas of similar shapes in gray on the CL images. The below pair of images should have a similar translation as the above pair:
When running this pair through the code below, the alignment is visually bad, and in fact the translation is in the wrong direction:
I have also been experimenting with filtering, edge detection and feature matching, using ORB for example, but the features detected are often not common between the pair of images and the matching doesn't seem to work very well
Is there anything I can do to improve my image registration algorithm to achieve what could be done fairly easily (but very slowly) by hand? Is there an alternative approach which may provide better results for what should be a small translation of images that are quite different in nature but include some correlatable elements?
import cv2
import numpy as np
import matplotlib
from matplotlib import pyplot as plt
def calculate_warp_translate(img1, img2, mask=None, init_warp_matrix=None, warp_mode=cv2.MOTION_TRANSLATION, number_of_iterations=1000, termination_eps=1e-15):
#Convert images to grayscale if necessary
if img1.ndim == 3:
img1 = cv2.cvtColor(img1,cv2.COLOR_BGR2GRAY)
if img2.ndim == 3:
img2 = cv2.cvtColor(img2,cv2.COLOR_BGR2GRAY)
# Find size of image1
sz = img1.shape
# Define 2x3 matrix and initialize the matrix to identity
if np.any(init_warp_matrix) == None:
warp_matrix = np.eye(2, 3, dtype=np.float32)
else:
warp_matrix = init_warp_matrix
# Define termination criteria
criteria = (cv2.TERM_CRITERIA_EPS | cv2.TERM_CRITERIA_COUNT, number_of_iterations, termination_eps)
# Run the ECC algorithm. The results are stored in warp_matrix.
cc, warp_matrix = cv2.findTransformECC(img1, img2, warp_matrix, warp_mode, criteria)
return warp_matrix
#image_name = 'image10_12_1.tif'
image_name = 'image10_13_1.tif'
img_BSE = cv2.imread('dataset1/BSE/' + image_name)
img_CL = cv2.imread('dataset1/CL/' + image_name)
warp_matrix = calculate_warp_translate(img_BSE, img_CL)
img_CL_aligned = cv2.warpAffine(img_CL, warp_matrix, (img_BSE.shape[1], img_BSE.shape[0]), flags=cv2.INTER_LINEAR + cv2.WARP_INVERSE_MAP)
plt.imshow(cv2.addWeighted(img_BSE, 0.5, img_CL, 0.5, 0), cmap='gray')
plt.title('Before alignment')
plt.show()
plt.imshow(cv2.addWeighted(img_BSE, 0.5, img_CL_aligned, 0.5, 0), cmap='gray')
plt.title('After alignment, warp matrix {}'.format(warp_matrix))
plt.show()
Images sourced from: Keulen, Nynke; Mette Olivarius; Nikolai Andrianov, 2022, "Scanning electron microscope images dataset for geothermal reservoir characterisation", https://doi.org/10.22008/FK2/5TWAZK, GEUS Dataverse, V2, http://creativecommons.org/licenses/by/4.0