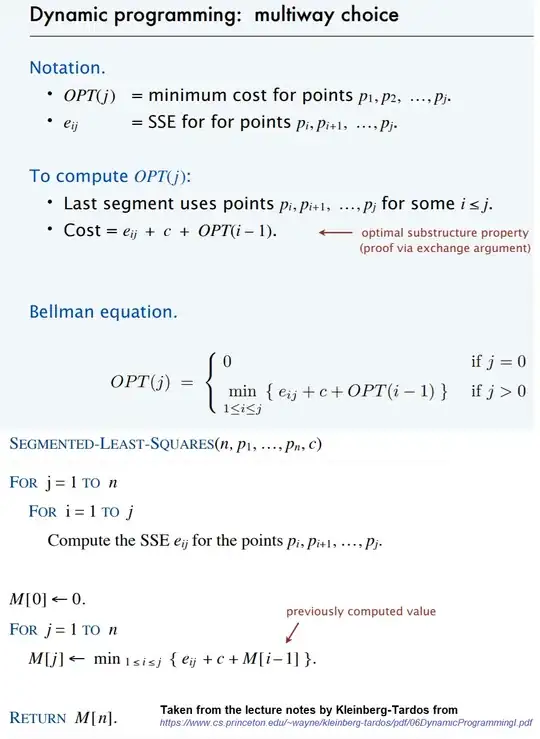
I have a ggplot function (ggproto) to generate a manhattan plot. The code works, but I can't get my function to correctly position the breaks and labels on the x-axis. So I would really appreciate help on: How I can get my function to plot the axis that I want. Is it possible within the stat_manhattan function or would it be better to code an extra scale_x_chr(or so) function? Many thanks in advance! P.S hope my question is clear.
This is the code for the function:
StatManhattan <- ggplot2::ggproto("StatManhattan",
ggplot2::Stat,
# set up parameters, e.g. unpack from list
setup_params = function(data, params) {
params
},
# Compute group is the most granular component to be called
compute_group = function(data, scales, params, col_chrom) {
message("My param has value ", scales)
data_1 <- data %>%
dplyr::arrange(chr, pos) %>%
dplyr::transmute(x = cumsum(as.numeric(pos)),
y,
chr)
data_2 <- data_1 %>%
dplyr::distinct(chr) %>%
dplyr::mutate(colour = rep(col_chrom, length.out = dplyr::n()))
# the final df
data_1 %>%
dplyr::left_join(data_2, by = "chr") %>%
as.data.frame()
},
required_aes = c("y", "pos", "chr"),
default_aes = aes(y = stat(y),
x = stat(x),
colour = stat(colour),
size = 0.2)
)
stat_manhattan <- function(mapping = NULL, data = NULL, geom = "point",
position = "identity", na.rm = FALSE, show.legend = FALSE,
inherit.aes = TRUE, col_chrom = c("magenta2", "grey60"),
...) {
ggplot2::layer(
stat = StatManhattan, data = data, mapping = mapping, geom = geom,
position = position, show.legend = show.legend, inherit.aes = inherit.aes,
params = list(na.rm = na.rm,
col_chrom = col_chrom,
...)
)
}
This can generate the desired plot, but the x-axis has continuous labels - which is not what I want.
# test data
test_data <- tibble(chr = rep(1:3, each = 1000),
bp = c(sort(sample.int(1e6, 1000)),
sort(sample.int(1e6, 1000)),
sort(sample.int(1e6, 1000))),
p = runif(3000, min=0, max=1))
# plotting the test data
ggplot(test_data)+aes(chr = chr, pos = bp, y =-log10(p))+stat_manhattan()
What I want to achieve is something like this, where the chromosome (chr) numbers are centered on the x-axis.
axisdf = test_data %>%
mutate(bp_cum = cumsum(as.numeric(bp))) %>%
group_by(chr) %>%
summarize(center=( max(bp_cum) + min(bp_cum) ) / 2 )
test_data %>%
mutate(bp_cum = cumsum(as.numeric(bp))) %>%
ggplot(., aes(x=bp_cum, y=-log10(p), colour = as.character(chr))) +
geom_point() +
# custom X axis:
scale_x_continuous(label = axisdf$chr, breaks= axisdf$center )