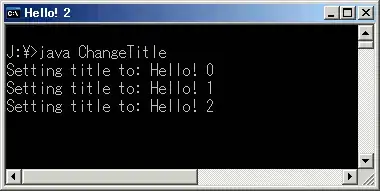
I am new to the Sjplot plot_model function. I am trying to plot a linear model, where the response variable is a proportion (min 0, max 1), yet when plotting this model the predicted values are plotted for negative values too.
The simple model I use;
sexpredictionadult <- lm(Propmort~Maletofemale,data=subset)
I try to correct this using +ylim, but this cuts part of the plot away.
plot_model(sexpredictionadult,type="slope",terms=c("Maletofemale"))
plot_model(sexpredictionadult,type="slope",terms=c("Maletofemale"))+ylim(c(0,1))
structure(list(Maletofemale = c(0.749999997916667, 0.0123456790047249,
0, NA, 0, 0, 0, 0.387499999515625, 0.3999999984, 0, 0.820512818408941,
0.47712418269469, 0, 0.461538457988166, 0, 5.8e+08, 6.6e+08,
0, 0, 0.19148936156632, 0.786885244611664, 0, 0.681415928902028,
2.38e+09, 0.249999999107143, 0, 0, 0.35999999928, 5.3e+08, NA,
0, 0, 0, 0.290540540344229, 0.34821428540338, NA, 0, 2.38e+09,
0, 0, 0, 0.195402298625974, 3.66666660555556, 1.30769229763314,
0, 0.363636361983471, 1.36842104903047, 0.312499998046875, 0,
0.315789472022161, 0.499999997222222, 0.545454542975207, 0.619047616099773,
0, 0.211864406600115, 0.866666660888889, 0.818181810743802, 7.3e+08,
0.499999998076923, 0.102564102301118, 8e+08, 0, 0.999999991666667,
0.405405405131483, 0, 1.06249999335938, 1.15789473074792, 0,
0, NA, NA, NA, NA, 1.02777777635031, 0, 2.85714281632653, NA,
0, 0, 0, NA, NA, 0, 0, 0, 0, 0.146245059230733, 0.52054794484894,
0.938931296993182, 0.958333329340278, 0, 0.999999993333333, 1.73e+09,
0.547619046315193, 0, 7.6e+08, NA, 0, NA, 0, 0, 0.365853657644259,
0, 3.4e+08, 0, 0, 0, NA, NA, NA, 0.307692305325444, 0.34999999825,
0.7368421033241, NA, NA, 0, 0, 0, 0, 0, 0, 0, 6e+08, 0, 0, 0,
0, 0, 1.08333332881944, 0, 0, NA, 0.50561797695998, 0.428571425510204,
0.666666661111111, 0, 0, 0, 0.77777777345679, NA, 0, 0, 0.8999999991,
0.424242422956841, NA, 0.901639342784198, 0.653846151331361,
4.8e+08, 0.99999999, 0.999999999363057, 0.821428568494898, 0.627272726702479,
0.411764703460208, 0.349999999125, NA, 7.5e+08, 0.555555549382716,
3.76923075956607, 0.232142856728316, 1.888888881893, 0.0487804876859012,
0, 0, 0, 0, 0.478260869218652, 0, 0, 0.999999994444444, 0, 0.652542372328354,
0, 0.655172412663496, 0.451612902983004, 0, 0, 0.636363630578513,
0, 0.266666665777778, NA, 2.1e+08, 5.2e+08, 0, 0.714285709183674,
1.15789473074792, 0, 0, NA, 0.404761903798186, 0, 1.06451612559834,
0, NA, 0.07999999968, 0.303030302112029, 0.116279069497025, NA,
NA, 0, 0, 0, NA, 0, NA, NA, 0, 0, 0, 0.622222220839506, 1.60439560263253,
0, 0.631578945706371, 0, 0, 0.487179485930309, 0, 0, 5.4999999828125,
1.94e+09, NA, NA, 0, 0, 0, NA, 3.09090906280992, 0.918367345064556,
NA, NA, 5.56e+09, 4.8e+09, 1.49999998928571, 0, 4.8e+08, 0, 0.285714283673469,
0, 0.256097560663296, 1.799999964, 0.562499996484375, 0.399999996,
0.102040816118284, 0.71739130278828, 0.142011834277511, 0.514285712816327,
0, 0.717948716107824), Propmort = c(0.6, 0.25, 1, 0.391304347826087,
0.0689655172413793, 0.133333333333333, 0.928571428571429, 0.380952380952381,
0.4375, 0.125, 0.0294117647058824, 0.258620689655172, 0, 0, 0.176470588235294,
0.32, 0.0689655172413793, 0.857142857142857, 0.333333333333333,
0.730769230769231, 0.521739130434783, 0.4, 0.461538461538462,
0, 0.75, 0.0909090909090909, 1, 0, 0.1, 0.0476190476190476, 0.625,
0.727272727272727, 0.375, 0.348484848484849, 0.26530612244898,
0.2, 0.111111111111111, 0, 0, 0, 0.368421052631579, 0.0555555555555556,
0, 0.181818181818182, 0, 0, 0, 0.666666666666667, 0.546511627906977,
0.6, 0.666666666666667, 0.857142857142857, 0.466666666666667,
0, 0.446808510638298, 0, 1, 0.384615384615385, 0, 0.142857142857143,
0.0967741935483871, 0, 0.833333333333333, 0, 0.518518518518518,
0, 0, 0.866666666666667, 1, 0.5, 0.2, 0.32258064516129, 0.588235294117647,
0.209302325581395, 0.3, 0.666666666666667, 0, 0.740740740740741,
0, 0, 0, 0.666666666666667, 0.166666666666667, 0.0384615384615385,
0, 0, 0, 0, 0, 0, 0.205882352941176, 0, 0, 0.411764705882353,
0, 0, 0, 0.0555555555555556, 0.535714285714286, 0.60655737704918,
0.418181818181818, 0, 0, 0.454545454545455, 0.242990654205607,
0.574074074074074, 0.75, 0, 0, 0, 0.666666666666667, 0.166666666666667,
0.1875, 0.771084337349398, 0.780821917808219, 0.888888888888889,
0, 0.32, 0.857142857142857, 0.516129032258065, 0.7, 0.277777777777778,
0, 0.775, 0.357142857142857, 0.714285714285714, 0, 0, 1, 0.785185185185185,
0, 0.75, 0.0326086956521739, 0.166666666666667, 0.428571428571429,
0.777777777777778, 0.736842105263158, 0.846153846153846, 0, 0.615384615384615,
0.75, 1, 0.315068493150685, 0.615384615384615, 0.333333333333333,
0.0625, 0.666666666666667, 0, 1, 0.0344827586206897, 0.428571428571429,
0.0444444444444444, 0, 0.4, 0.676470588235294, 0.0526315789473684,
0, 0.136986301369863, 0, 0.0512820512820513, 0.5, 1, 0.805555555555556,
0.6875, 0, 0, 0.0909090909090909, 0.458333333333333, 0.25, 0,
0.539473684210526, 0, 0, 0.150943396226415, 0, 0.666666666666667,
0, 0.5, 0.75, 0.386363636363636, 0, 0.0666666666666667, 0.0465116279069767,
0, 0.181818181818182, 0, 0.0909090909090909, 0, 0, 0, 0.0526315789473684,
0.1, 0.0833333333333333, 0.545454545454545, 0.764705882352941,
0.133333333333333, 0.75, 0, 0.5, 0.9, 0.230769230769231, 0.454545454545455,
0.397260273972603, 1, 0.722222222222222, 0.142857142857143, 0.117647058823529,
0.181818181818182, 0.292682926829268, 0, 0.607142857142857, 0.307692307692308,
0.363636363636364, 0.814814814814815, 0.444444444444444, 0.625,
0, 0.0289855072463768, 0.483516483516484, 0.702702702702703,
0.75, 0.19047619047619, 0, 0.181818181818182, 0.806451612903226,
0, 0.75, 0, 0.260869565217391, 0, 0.0175438596491228, 0, 0, 0,
1, 0.5, 0.173913043478261, 0.488372093023256, 0, 0.666666666666667,
0, 0.666666666666667, 0, 0.411042944785276, 0.176470588235294,
0.894736842105263, 0)), row.names = c(2L, 3L, 7L, 9L, 12L, 13L,
16L, 21L, 22L, 23L, 24L, 25L, 28L, 29L, 30L, 31L, 32L, 37L, 39L,
40L, 41L, 43L, 45L, 48L, 50L, 51L, 53L, 54L, 55L, 57L, 59L, 60L,
61L, 63L, 67L, 70L, 71L, 73L, 74L, 75L, 77L, 78L, 79L, 80L, 81L,
82L, 83L, 84L, 85L, 86L, 87L, 88L, 89L, 90L, 91L, 93L, 94L, 95L,
96L, 98L, 100L, 103L, 104L, 108L, 109L, 110L, 111L, 113L, 114L,
115L, 116L, 117L, 118L, 120L, 121L, 122L, 123L, 124L, 126L, 128L,
129L, 131L, 133L, 140L, 144L, 145L, 148L, 149L, 150L, 151L, 152L,
154L, 157L, 159L, 160L, 161L, 162L, 163L, 164L, 165L, 166L, 167L,
168L, 170L, 171L, 173L, 174L, 177L, 178L, 179L, 180L, 182L, 184L,
185L, 186L, 190L, 191L, 193L, 198L, 200L, 203L, 204L, 205L, 207L,
209L, 210L, 212L, 213L, 214L, 215L, 220L, 221L, 222L, 223L, 224L,
225L, 226L, 227L, 229L, 230L, 231L, 232L, 234L, 235L, 236L, 238L,
239L, 240L, 241L, 244L, 245L, 246L, 247L, 249L, 251L, 252L, 253L,
254L, 255L, 256L, 260L, 262L, 263L, 264L, 265L, 266L, 267L, 269L,
271L, 272L, 277L, 278L, 280L, 281L, 282L, 283L, 284L, 285L, 291L,
294L, 295L, 296L, 297L, 303L, 305L, 306L, 307L, 308L, 309L, 312L,
316L, 317L, 319L, 320L, 321L, 322L, 324L, 325L, 329L, 330L, 331L,
333L, 334L, 335L, 336L, 339L, 343L, 345L, 349L, 350L, 352L, 355L,
356L, 357L, 359L, 361L, 362L, 364L, 366L, 367L, 368L, 369L, 370L,
372L, 376L, 377L, 378L, 380L, 382L, 386L, 387L, 391L, 393L, 394L,
395L, 396L, 397L, 399L, 400L, 403L, 404L, 407L, 409L, 410L, 412L,
413L, 416L), class = "data.frame")
dataset: