I have written a function which evaluates the Laplace approximate marginal likelihood of complex mixed models. The implementation uses sparse matrices with Eigen/RcppEigen, and the C++ autodiff library. It is available by installing the development version of the galamm package using the code in the line below. The installation requires compilation of C++ code, but the package passes R CMD check, so it should be easy to install for anyone with R set up for building packages.
remotes::install_github("LCBC-UiO/galamm",
ref = "7b0b521bb6a9aeb2c93da39dc7a07fef61d81211")
In the package I have a function marginal_likelihood which evaluates the marginal likelihood of the model at given values of the parameters. The function returns a list containing two elements: (1) the deviance of the model, and (2) the gradient with respect to the parameters.
Although the function is intended for latent variable models used in psychometrics, for the purpose of this question it is sufficient to use a linear mixed model. In the code directly below, I confirm that the deviance is identical to the computed by lme4::lmer, using an example problem for lme4. It should also be clear the marginal_likelihood is intended to be a low-level function, not directly exposed to users.
library(lme4)
#> Loading required package: Matrix
library(galamm)
# Fit a linear mixed model using lme4's modular fitting functions
lmod <- lFormula(Reaction ~ Days + (Days|Subject), sleepstudy, REML = FALSE)
devfun <- do.call(mkLmerDevfun, lmod)
opt <- optimizeLmer(devfun)
mod1 <- mkMerMod(environment(devfun), opt, lmod$reTrms, fr = lmod$fr)
# Confirm that galamm::marginal_likelihood returns the correct value at the
# maximum likelihood estimates
mod2 <- marginal_likelihood(
y = sleepstudy$Reaction,
trials = 0, # not used here
X = lmod$X,
Zt = lmod$reTrms$Zt,
Lambdat = lmod$reTrms$Lambdat,
beta = fixef(mod1),
theta = getME(mod1, "theta"),
theta_mapping = lmod$reTrms$Lind - 1L,
lambda = numeric(), # factor loadings, not used here
lambda_mapping_X = integer(),
lambda_mapping_Zt = integer(),
family = "gaussian"
)
-2 * logLik(mod1)
#> 'log Lik.' 1751.939 (df=6)
mod2$deviance
#> [1] 1751.939
# marginal_likelihood also returns the gradient wrt theta, beta, and lambda
mod2
#> $deviance
#> [1] 1751.939
#>
#> $gradient
#> [1] -1.392026e-03 -1.483929e-03 -1.407926e-03 1.327937e-14 -1.724646e-13
Created on 2022-08-18 with reprex v2.0.2
What I now want to do is use the two elements of the list returned by marginal_likelihood, i.e., $deviance and $gradient, to find marginal maximum likelihood estimates using optim(). Since the gradient is computed using automatic differentiation in C++, it is wasteful to compute this in a second run of the function. Hence, I have implemented the following "hack", which works upto some point:
library(lme4)
#> Loading required package: Matrix
library(galamm)
lmod <- lFormula(Reaction ~ Days + (Days|Subject), sleepstudy, REML = FALSE)
theta_inds <- seq(from = 1, to = length(lmod$reTrms$theta))
beta_inds <- seq(from = max(theta_inds) + 1, length.out = ncol(lmod$X))
bounds <- c(lmod$reTrms$lower, rep(-Inf, length(beta_inds)))
optfun <- function(par){
ml <- marginal_likelihood(
y = sleepstudy$Reaction,
trials = 0, # not used here
X = lmod$X,
Zt = lmod$reTrms$Zt,
Lambdat = lmod$reTrms$Lambdat,
beta = par[beta_inds],
theta = par[theta_inds],
theta_mapping = lmod$reTrms$Lind - 1L,
lambda = numeric(), # factor loadings, not used here
lambda_mapping_X = integer(),
lambda_mapping_Zt = integer(),
family = "gaussian"
)
# Update gradient globally
grad <<- ml$gradient
ml$deviance
}
# Simply take the gradient from the global environment
gradfun <- function(par){
grad
}
# Set gradient to NULL
grad <- NULL
par_init <- c(lmod$reTrms$theta, rep(1, length(beta_inds)))
opt <- optim(par_init, fn = optfun, gr = gradfun, lower = bounds,
method = "L-BFGS-B")
opt
#> $par
#> [1] 0.92992479 0.01811702 0.22274704 251.40024419 10.46528199
#>
#> $value
#> [1] 1751.939
#>
#> $counts
#> function gradient
#> 116 116
#>
#> $convergence
#> [1] 0
#>
#> $message
#> [1] "CONVERGENCE: REL_REDUCTION_OF_F <= FACTR*EPSMCH"
# Confirm that estimates are identical to lme4:
mod1 <- lmer(Reaction ~ Days + (Days|Subject), sleepstudy, REML = FALSE)
plot(getME(mod1, "theta"), opt$par[theta_inds]); abline(0, 1)
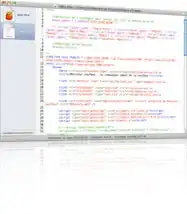
plot(fixef(mod1), opt$par[beta_inds]); abline(0, 1)

Created on 2022-08-18 with reprex v2.0.2
It does converge to the right solution, but my has several problems, including:
- It uses global assignment to update the gradient.
- It is dependent on the fact the L-BFGS-B compute the gradient directly after evaluating the objective, for any given parameter value. For this reason, if I set
hessian = TRUEin the argument tooptim(), I get a matrix of zeros, since when evaluating the Hessian at the end of the iterations, this order does no longer hold.
My question thus is: Is there a nicer way of minimizing a function using optim() or other alternatives like optimx() or optimr(), when the function being optimized returns both the function value AND the gradient at the given parameters?