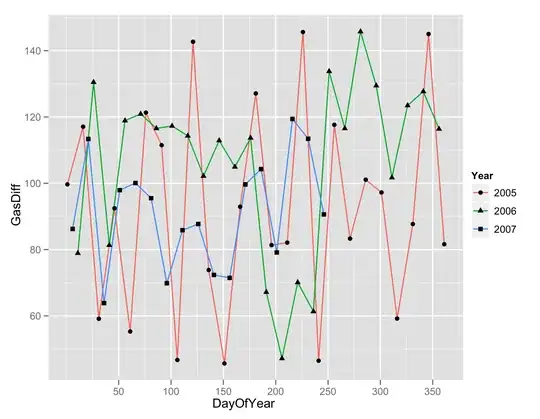
After I made a ggplot of my data
I'd like to adjust each curve with a function. I first adjust with nls function, but for the dark blue one, it doesn't work, and it's obvious when we see the curve shape.
So I want to make a logistic function because of the sigmoid shape, but it doesn't work again.
Here is my code, with my starting values:
start.values <- list(gm=0.6, k=0.8)
nls4 = nls(growth_rate ~ gm/(1+(exp(-0.8*(quantity-k)))), data = pr28S, start=start.values)
gm is the maximum value of the curve, k is the x value of the sigmoid's midpoint and 0.8 is the logistic growth rate. quantity is my x axis and growth_rate my y axis.
I tried different starting values and change the 0.8 value between 0.5 and 2 but it never work. I have different errors like :
'Error in nls(growth_rate ~ gm/(1 + (exp(-0.8 * (quantity - k)))), data = pr28S, : the step 0.000488281 is lower than'minFactor' of 0.000976562'
or also: 'singular gradient'
How can I adjust my curve pls ?
EDIT : Here is my data for the 28S curve :
structure(list(name = c("J4_S01AC", "J4_S01CC", "J4_S01EC", "J4_S01FC",
"J4_S03BC", "J4_S03CC", "J4_S03DC", "J4_S03EC", "J4_S03FC", "J4_S06AC",
"J4_S06DC", "J4_S06EC", "J4_S06FC", "J4_S06KC", "J4_S06MC", "J4_S06NC",
"J4_S09AC", "J4_S09BC", "J4_S09CC", "J4_S09DC", "J4_S03AM", "J4_S03BM",
"J4_S03CM", "J4_S15AM", "J4_S15BM", "J4_S15CM", "J4_S15DM", "J4_S01AAC"
), day = c("J4", "J4", "J4", "J4", "J4", "J4", "J4", "J4", "J4",
"J4", "J4", "J4", "J4", "J4", "J4", "J4", "J4", "J4", "J4", "J4",
"J4", "J4", "J4", "J4", "J4", "J4", "J4", "J4"), quality = c("S",
"S", "S", "S", "S", "S", "S", "S", "S", "S", "S", "S", "S", "S",
"S", "S", "S", "S", "S", "S", "S", "S", "S", "S", "S", "S", "S",
"S"), quantity = c(0.1, 0.1, 0.1, 0.1, 0.3, 0.3, 0.3, 0.3, 0.3,
0.6, 0.6, 0.6, 0.6, 0.6, 0.6, 0.6, 0.9, 0.9, 0.9, 0.9, 0.3, 0.3,
0.3, 1.5, 1.5, 1.5, 1.5, 0.1), qual_quant = c("S01", "S01", "S01",
"S01", "S03", "S03", "S03", "S03", "S03", "S06", "S06", "S06",
"S06", "S06", "S06", "S06", "S09", "S09", "S09", "S09", "S03",
"S03", "S03", "S15", "S15", "S15", "S15", "S01"), temperature = c(28,
28, 28, 28, 28, 28, 28, 28, 28, 28, 28, 28, 28, 28, 28, 28, 28,
28, 28, 28, 28, 28, 28, 28, 28, 28, 28, 28), time = c(101, 101,
101, 101, 101, 101, 101, 101, 101, 101, 101, 101, 101, 101, 101,
101, 102, 102, 102, 102, 107.5, 107.5, 107.5, 107.5, 107.5, 107.5,
107.5, 109), size = c(1344.366, 1291.962, 1304.811, 1128.264,
1286.151, 1358.421, 1396.671, 1353.076, 1505.565, 1297.17, 0,
1243.029, 1323.85, 1368.364, 1506.396, 0, 1663.735, 1632.28,
2115.303, 1921.46, 1506.581, 1501.196, 1370.99, 1870.489, 1941.425,
1942.186, 1827.395, 1336.588), weight = c(11, 16, 10, 10, 12,
12, 13, 16, 14, 12, 11, 12, 10, 10, 15, 25, 46, 35, 66, 46, 20,
16, 15, 49, 73, 63, 60, 11), growth_rate = c(0.0378568479840844, 0.126892202895244,
0.0152090656484838, 0.0152090656484838, 0.0585326533474545, 0.0585326533474545,
0.0775525505364441, 0.126892202895244, 0.0951622248242906, 0.0585326533474545,
0.0378568479840844, 0.0585326533474545, 0.0152090656484838, 0.0152090656484838,
0.111556439370444, 0.232939774941222, 0.374129156018743, 0.309825356331568,
0.459072793066665, 0.374129156018743, 0.169037347727828, 0.119219651092275,
0.104811166292231, 0.369092608582107, 0.458090402952369, 0.425199566970549,
0.414306966296783, 0.0350783637283718), color = c("28S", "28S",
"28S", "28S", "28S", "28S", "28S", "28S", "28S", "28S", "28S",
"28S", "28S", "28S", "28S", "28S", "28S", "28S", "28S", "28S",
"28S", "28S", "28S", "28S", "28S", "28S", "28S", "28S")), row.names = c(NA,
-28L), class = c("tbl_df", "tbl", "data.frame"))