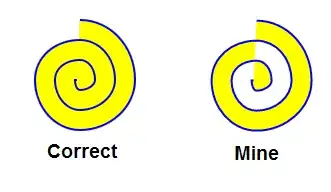
I have a question where I don´t know how to start this. We did a scRNA sequencing experiment and I now have an AnnData dataset. I already know a lot about this dataset mainly by using scanpy library and I would like to "finalise" the analysis by extracting genes that have a specific RNA sequence in the 3'UTR. Unfortunately I have no idea how to approach this since I am no bioinformatician and couldn´t find a tutorial to do this. Can someone please help me with this problem?
Thanks in advance!