I want to draw the lattice of subgroups up to a finite subgroup index of an infinite, discrete space group with a graph drawing tool such as yEd, GraphViz, NetworkX, ...
An Example input file
would be following graphml file for the two-dimensional space group p4gm up to index 8 (generated by self-written code in gap):
<?xml version='1.0' encoding='UTF-8'?>
<graphml
xmlns='http://graphml.graphdrawing.org/xmlns'
xmlns:xsi='http://www.w3.org/2001/XMLSchema-instance'
xsi:schemaLocation='http://graphml.graphdrawing.org/xmlns
http://graphml.graphdrawing.org/xmlns/1.0/graphml.xsd'>
<key id='idx' for='node' attr.name='index' attr.type='int' />
<key id='r' for='node' attr.name='radius' attr.type='double' />
<key id='idx' for='edge' attr.name='index' attr.type='int' />
<graph id='G' edgedefault='directed'>
<node id='01'> <data key='idx'>1</data> <data key='r'>0.</data> </node>
<node id='02'> <data key='idx'>2</data> <data key='r'>0.33333333333333337</data> </node>
<node id='03'> <data key='idx'>2</data> <data key='r'>0.33333333333333337</data> </node>
<node id='04'> <data key='idx'>2</data> <data key='r'>0.33333333333333337</data> </node>
<node id='05'> <data key='idx'>4</data> <data key='r'>0.66666666666666674</data> </node>
<node id='06'> <data key='idx'>4</data> <data key='r'>0.66666666666666674</data> </node>
<node id='07'> <data key='idx'>6</data> <data key='r'>0.8616541669070521</data> </node>
<node id='08'> <data key='idx'>4</data> <data key='r'>0.66666666666666674</data> </node>
<node id='09'> <data key='idx'>4</data> <data key='r'>0.66666666666666674</data> </node>
<node id='10'> <data key='idx'>4</data> <data key='r'>0.66666666666666674</data> </node>
<node id='11'> <data key='idx'>4</data> <data key='r'>0.66666666666666674</data> </node>
<node id='12'> <data key='idx'>4</data> <data key='r'>0.66666666666666674</data> </node>
<node id='13'> <data key='idx'>6</data> <data key='r'>0.8616541669070521</data> </node>
<node id='14'> <data key='idx'>8</data> <data key='r'>1.</data> </node>
<node id='15'> <data key='idx'>8</data> <data key='r'>1.</data> </node>
<node id='16'> <data key='idx'>8</data> <data key='r'>1.</data> </node>
<node id='17'> <data key='idx'>8</data> <data key='r'>1.</data> </node>
<node id='18'> <data key='idx'>8</data> <data key='r'>1.</data> </node>
<node id='19'> <data key='idx'>8</data> <data key='r'>1.</data> </node>
<node id='20'> <data key='idx'>8</data> <data key='r'>1.</data> </node>
<node id='21'> <data key='idx'>8</data> <data key='r'>1.</data> </node>
<node id='22'> <data key='idx'>8</data> <data key='r'>1.</data> </node>
<node id='23'> <data key='idx'>8</data> <data key='r'>1.</data> </node>
<node id='24'> <data key='idx'>8</data> <data key='r'>1.</data> </node>
<node id='25'> <data key='idx'>8</data> <data key='r'>1.</data> </node>
<node id='26'> <data key='idx'>8</data> <data key='r'>1.</data> </node>
<edge id='e01' target='02' source='01'> <data key='idx'>2</data> </edge>
<edge id='e02' target='03' source='01'> <data key='idx'>2</data> </edge>
<edge id='e03' target='04' source='01'> <data key='idx'>2</data> </edge>
<edge id='e04' target='05' source='02'> <data key='idx'>2</data> </edge>
<edge id='e05' target='06' source='02'> <data key='idx'>2</data> </edge>
<edge id='e06' target='07' source='02'> <data key='idx'>3</data> </edge>
<edge id='e07' target='06' source='03'> <data key='idx'>2</data> </edge>
<edge id='e08' target='08' source='03'> <data key='idx'>2</data> </edge>
<edge id='e09' target='06' source='04'> <data key='idx'>2</data> </edge>
<edge id='e10' target='10' source='04'> <data key='idx'>2</data> </edge>
<edge id='e11' target='11' source='04'> <data key='idx'>2</data> </edge>
<edge id='e12' target='09' source='04'> <data key='idx'>2</data> </edge>
<edge id='e13' target='12' source='04'> <data key='idx'>2</data> </edge>
<edge id='e14' target='13' source='04'> <data key='idx'>3</data> </edge>
<edge id='e15' target='14' source='05'> <data key='idx'>2</data> </edge>
<edge id='e16' target='15' source='05'> <data key='idx'>2</data> </edge>
<edge id='e17' target='14' source='06'> <data key='idx'>2</data> </edge>
<edge id='e18' target='16' source='06'> <data key='idx'>2</data> </edge>
<edge id='e19' target='17' source='06'> <data key='idx'>2</data> </edge>
<edge id='e20' target='18' source='06'> <data key='idx'>2</data> </edge>
<edge id='e21' target='16' source='08'> <data key='idx'>2</data> </edge>
<edge id='e22' target='19' source='08'> <data key='idx'>2</data> </edge>
<edge id='e23' target='18' source='09'> <data key='idx'>2</data> </edge>
<edge id='e24' target='20' source='09'> <data key='idx'>2</data> </edge>
<edge id='e25' target='14' source='10'> <data key='idx'>2</data> </edge>
<edge id='e26' target='16' source='11'> <data key='idx'>2</data> </edge>
<edge id='e27' target='21' source='11'> <data key='idx'>2</data> </edge>
<edge id='e28' target='22' source='11'> <data key='idx'>2</data> </edge>
<edge id='e29' target='23' source='11'> <data key='idx'>2</data> </edge>
<edge id='e30' target='18' source='12'> <data key='idx'>2</data> </edge>
<edge id='e31' target='21' source='12'> <data key='idx'>2</data> </edge>
<edge id='e32' target='24' source='12'> <data key='idx'>2</data> </edge>
<edge id='e33' target='25' source='12'> <data key='idx'>2</data> </edge>
<edge id='e34' target='26' source='12'> <data key='idx'>2</data> </edge>
</graph>
</graphml>
I have anonymous-ed the data to focus on the graph drawing.
I am looking for a graph drawing tool which can layout the nodes on a radial layout, similar to a radial tree but can draw non-straight edges to avoid edge-node crossings. Edge-edge crossing are fine. Ideally however, a viewer can follow each edge from source to target node.
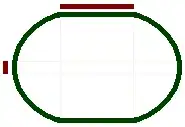
yEd (3.22)
provides a radial layout which can draw edges as arcs or curved to avoid edge-node crossings:
However, the nodes are placed on the same concentric circle based on the shortest distance to the center, measured by number of traversed edges.
But I want to place the nodes based on their subgroup index (to be precise the logarithm of the index). In the above picture the nodes with index 6 are on the same circle as the nodes with index 4 which is not what I want.
NetworkX (2.8.4)
has the shell layout which allows you to assign manually the nodes to the shells
import networkx as nx
import matplotlib.pyplot as plt
from math import log
G = nx.read_graphml("ITC_2_012_idx8.graphml")
indices = set([idx for n, idx in G.nodes.data('index')])
radii = [log(idx)/log(max(indices)) for n, idx in G.nodes.data('index')]
shells = [[n for n, idx in G.nodes.data('index') if idx == x] for x in indices]
pos = nx.shell_layout(G, shells)
plt.box(False) # remove box
nx.draw_networkx(G, pos,
node_color="white",
node_size=500,
edgecolors="black",
labels={n: idx for n, idx in G.nodes.data('index')},
)
However, NetworkX draws only straight edges.
Can GraphViz or another graph drawing tool do what I want?
I have started to create an own layouting and edge routing algorithm which results in following style of drawing:
However, this is unfinished and becomes a never ending story. So I am hoping that I have overlooked a tool which can give automatically the desired radial layout and suitable edge routes. yEd is the closest tool I have come by (see the first picture of this question).