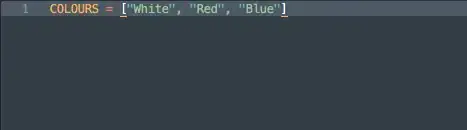
I have the following log-likelihood from my model which i am trying to write as a function in R.
My issue come as i dont know how to write theta in terms of the the function. I have had a couple of attempts at this as shown below, any tips/advice on if these are close to being correct could be appreciated.
function with theta written as theta
#my likelihood function
mylikelihood = function(beta) {
#log-likelihood
result = sum(log(dengue$cases + theta + 1 / dengue$cases)) +
sum(theta*log(theta / theta + exp(beta[1]+beta[2]*dengue$time))) +
sum(theta * log(exp(beta[1]+beta[2]*dengue$time / dengue$cases + exp(beta[1]+beta[2]*dengue$time))))
#return negative log-likelihood
return(-result)
}
my next attempt with thetas replaced with Xi from my dataset, which here is (dengue$time)
#my likelihood function attempt 2
mylikelihood = function(beta) {
#log-likelihood
result = sum((log(dengue$Cases + dengue$Time + 1 / dengue$Cases))) +
sum(dengue$Time*log(dengue$time / dengue$Time + exp(beta[1]+beta[2]*dengue$Time))) +
sum(dengue$Cases * log(exp(beta[1]+beta[2]*dengue$Time / dengue$Cases +
exp(beta[1]+beta[2]*dengue$Time))))
#return negative log-likelihood
return(-result)
}
data
head(dengue)
Cases Week Time
1 148 36 1
2 275 37 2
3 205 38 3
4 133 39 4
5 123 40 5
6 138 41 6
Are either of these close to being correct, and if not where am I going wrong?
Updated into about where the log-likelihood comes from;
The model;
Negative Binomial distribution with mean µ and dispersion parameter θ has pmf;