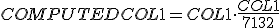Following this answer I am trying to plot a seaborn histplot with the pdf I get from scipy.stats. The problem is that the plotted pdf is catastrophically out of scale (example below):
from scipy import stats
import matplotlib.pyplot as plt
import seaborn as sns
params = (20, -1500, 8000)
sample = stats.invgauss.rvs(size=1000, *params)
fig, ax = plt.subplots()
sns.histplot(sample, log_scale=True, kde=False, stat='density')
x0, x1 = ax.get_xlim()
x_pdf = np.exp(np.linspace(np.log(x0),np.log(x1),500))
fitted_params = stats.invgauss.fit(sample)
y_pdf = stats.invgauss.pdf(x_pdf, *fitted_params)
y_cdf = stats.invgauss.cdf(x_pdf, *fitted_params)
ax.plot(x_pdf, y_pdf, 'r', lw=2)
plt.savefig('Pdf_check.png')
On the other hand, cdf prints fine and is consistent with what I would expect:
# same as above
ax.plot(x_pdf, y_cdf, 'r', lw=2)
plt.savefig('Cdf_check.png')