I am checking my data for normality and I have created boxplots - boxplot(), histograms - hist(), and ggqqplots() (using the ggpubr package, which is aligned with ggplot). The data shows measurements of acoustic parameters within spectrograms using Raven Software.
For this project, I need to visually represent my results neatly and I want to use plot_grid() in the cowplot Package to align my plots in 9 columns and 3 rows. I have produced 9 boxplots, 9 histograms, and 9 ggqqplot() objects.
When I attempted to align my plots using the plot_grid() function, the qqggplot() printed but the hist() and boxplots() objects did not (see diagram 1). I learned that I need to convert my hist() and boxplot() objects into grob objects.
I found a website (below) that stated could use as.glob() or as.ggplot() to convert my diagrams. I used the information from this link below, although, unsuccessfully.
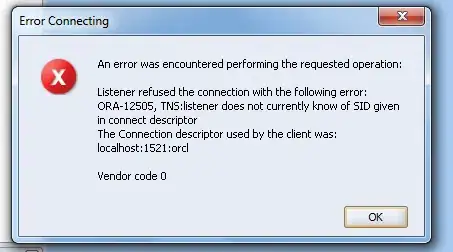
After I attempted to convert my hist() and boxplot() objects to grob object (so they will analogously be ggplot objects for the plot_grid() command) using the as.grob() function, I got error message 2 (see below).
R Code
#Install library packages
library("ggpubr")
library("grid")
library("ggplotify")
library(ggplot2)
library(cowplot)
#Box plots
Low_Freq_Box<-boxplot(main_path$Low.Freq..Hz.)
High_Freq_Box<-boxplot(main_path$High.Freq..Hz.)
Peak_Freq_Box<-boxplot(main_path$Peak.Freq..Hz.)
Delta_Freq_Box<-boxplot(main_path$Delta.Freq..Hz.)
Peak_Time_Box<-boxplot(main_path$ Peak.Time..s.)
Delta_Time_Box<-boxplot(main_path$Delta.Time..s.)
Center_Freq_Box<-boxplot(main_path$Center.Freq..Hz.)
Start_Freq_Box<-boxplot(main_path$Start.Freq..Hz.)
End_Freq_Box<-boxplot(main_path$End.Freq..Hz.)
#Histograms
Low_Freq_hist<-hist(main_path$Low.Freq..Hz.)
High_Freq_hist<-hist(main_path$High.Freq..Hz.)
Peak_Freq_hist<-hist(main_path$Peak.Freq..Hz.)
Delta_Freq_hist<-hist(main_path$Delta.Freq..Hz.)
Peak_Time_hist<-hist(main_path$Peak.Time..s.)
Delta_Time_hist<-hist(main_path$Delta.Time..s.)
Center_Freq_hist<-hist(main_path$Center.Freq..Hz.)
Start_Freq_hist<-hist(main_path$Start.Freq..Hz.)
End_Freq_hist<-hist(main_path$End.Freq..Hz.)
#QQ Plots
Low_Freq_qqplot<-ggqqplot(main_path$Low.Freq..Hz.)
High_Freq_qqplot<-ggqqplot(main_path$High.Freq..Hz.)
Peak_Freq_qqplot<-ggqqplot(main_path$Peak.Freq..Hz.)
Delta_Freq_qqplot<-ggqqplot(main_path$Delta.Freq..Hz.)
Delta_Time_qqplot<-ggqqplot(main_path$Delta.Time..s.)
Peak_Time_qqplot<-ggqqplot(main_path$Peak.Time..s.)
Center_Freq_qqplot<-ggqqplot(main_path$Center.Freq..Hz.)
Start_Freq_qqplot<-ggqqplot(main_path$Start.Freq..Hz.)
End_Freq_qqplot<-ggqqplot(main_path$End.Freq..Hz.)
Error Message 1
#Plot the box plots, histograms, and qqplots on the same grid
plot_grid(Low_Freq_Box, High_Freq_Box, Peak_Freq_Box, Delta_Freq_Box, Peak_Time_Box, Delta_Time_Box, Center_Freq_Box, Start_Freq_Box, End_Freq_Box,
Low_Freq_hist, High_Freq_hist, Peak_Freq_hist, Delta_Freq_hist, Peak_Time_hist, Delta_Time_hist, Center_Freq_hist, Start_Freq_hist, End_Freq_hist,
Low_Freq_qqplot, High_Freq_qqplot, Peak_Freq_qqplot, Delta_Freq_qqplot, Delta_Time_qqplot, Peak_Time_qqplot, Center_Freq_qqplot, Start_Freq_qqplot, End_Freq_qqplot,
labels=c("1", "2", "3", "4", "5", "6", "7", "8", "9", "10", "11", "12", "13", "14', 15", "16", "17", "18", "19", "20", "21", "22", "23", "24", "25", "26", "27"),
ncol=9, nrow=3)
#Warning Messages
plot_grid(Low_Freq_Box, High_Freq_Box, Peak_Freq_Box, Delta_Freq_Box, Peak_Time_Box, Delta_Time_Box, Center_Freq_Box, Start_Freq_Box, End_Freq_Box,
+ Low_Freq_hist, High_Freq_hist, Peak_Freq_hist, Delta_Freq_hist, Peak_Time_hist, Delta_Time_hist, Center_Freq_hist, Start_Freq_hist, End_Freq_hist,
+ Low_Freq_qqplot, High_Freq_qqplot, Peak_Freq_qqplot, Delta_Freq_qqplot, Delta_Time_qqplot, Peak_Time_qqplot, Center_Freq_qqplot, Start_Freq_qqplot, End_Freq_qqplot,
+ labels=c("1", "2", "3", "4", "5", "6", "7", "8", "9", "10", "11", "12", "13", "14", "15", "16", "17", "18", "19", "20", "21", "22", "23", "24", "25", "26", "27"),
+ ncol=9, nrow=3,
+ align="hv",
+ label_size = 12)
There were 20 warnings (use warnings() to see them)
Diagram 1