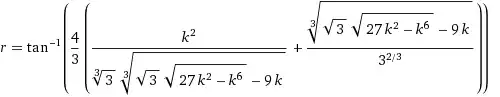I'm having a frustrating problem that I can't reproduce (I wish I could). I've generated dendrograms with three ecological datasets, using the same code but unique objects for each. Each leaf in the dendrograms is a survey plot, with species presence/abundance driving the clustering.
I cut the dendrogram into 3 groups, and color code each group. This works for fine for all three datasets when clustering using Euclidean distance, and for two of my datasets when using Bray-Curtis distance. However: the third dataset clusters two leaves when using Bray-Curtis, and forces the color code to recycle, creating k = 4 groups despite specifying k = 3.
My question is: why would two leaves (plots) be forced into their own 'cluster,' and force the dendrogram to have 4 clusters when I've specified k = 3 groups?
I've pasted below an example of the code, and images of the "correct" and "wrong" dendrograms. Curious if anyone has any troubleshooting suggestions, since I can't offer code that reproduces this error. TIA.
I've tried:
- removing the custom color value (no effect, still get 4 clusters when k = 3).
- adding a cutree argument to the 'dend' object, but this produces error 'Error in stats::cutree(tree, k = k, h = h, ...) : the 'height' component of 'tree' is not sorted (increasingly)'
Example code (same format with unique objects used for each dendrogram figure). Please access csv from https://drive.google.com/file/d/12eXIXVuHTu4BLGxcGu18bqhT85ZOHkNW/view?usp=sharing. See file clusterdata.csv for the troublesome dataset. Colnames are species; rows are plot ID; values are cover class bins (0 = absent, 1 = < 25%, 2 = 25-50%, etc.)
#library(dendextend)
d <- read.csv("clusterdata.csv")
dend <- d %>%
vegdist(method = "bray") %>%
hclust(method = "ward.D") %>%
# cutree(h = 3) %>%
as.dendrogram()
mycol <- c("#009E73", "#0072B2", "#E69F00")
dend.plot <- as.dendrogram(dend) %>%
set("branches_lwd", 2) %>% # Branches line width
set("branches_k_color", mycol, k = 3) %>% # Color branches by groups
set("labels_cex", 0.5) # Change label size
plot(dend.plot, ylab = "Bray-Curtis Distance", main = "why would clusters be different?")