I have read bunch of dask examples from either someone's GitHub code or the dask issues. But still have a problem of using Scipy interpolation with Dask parallel computing and hoping someone here can help me to solve it.
I actually have issue in how to expand each partition boundary. Please see my description below, and let me know if you cannot understand it.
- My data is unstructured and cannot using array
- My interpolation code is running, but there are some strange points occurring, I bet that is because of the edge effect. For example
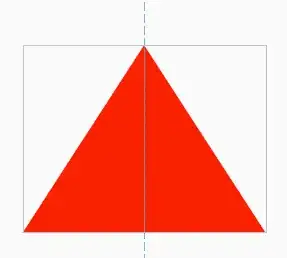 The left panel is the output from "dask" linearNDInterpolator, while the middle panel is the original dataset and the right panel is using linearNDInterpolator directly.
The left panel is the output from "dask" linearNDInterpolator, while the middle panel is the original dataset and the right panel is using linearNDInterpolator directly.
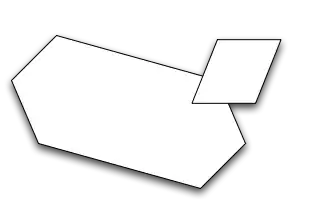 The left panel is a scatter plot that is linear interpolated from original dataset, while the right hand side one is using dask linearinterpolation by dask.dataframe [parallel].
The left panel is a scatter plot that is linear interpolated from original dataset, while the right hand side one is using dask linearinterpolation by dask.dataframe [parallel].
You can clearly see that the parallel computing results has no clear shape, and may possible see some strange points within the map.
Here is my code 01: Using dask.array.
def lNDIwrap(src_lon, src_lat, src_elv, tag_lon, tag_lat):
return lNDI(list(zip(src_lon, src_lat)), src_elv)(tag_lon, tag_lat)
n_splits=96
#--- topodf is the topography dataset [pandas].
#--- df is the python h3 generated hexagon grid by topodf in resolution-12
dsrc = dd.from_pandas(topodf, npartitions=n_splits)
dtag = dd.from_pandas(df, npartitions=n_splits)
#--- Output the chunking dask array
slon,slat,data = dsrc.to_dask_array(lengths=True).T
tlon,tlat = dtag[['lon','lat']].to_dask_array(lengths=True).T
#--- Using **dask delayed** function to pass each partition into functions [lNDIwrap]
gd_chunked_lNDI = [delayed(lNDIwrap)(x1, y1, newarr, xx, yy) for x1, y1, newarr, xx, yy in \
zip(slon.to_delayed().flatten(),
slat.to_delayed().flatten(),
data.to_delayed().flatten(),
tlon.to_delayed().flatten(),
tlat.to_delayed().flatten())]
#--- Using **dask delayed** function to concat all partitions into one np.array
gd_lNDI = delayed(da.concatenate)(gd_chunked_lNDI, axis=0)
results_lNDI = np.array(gd_lNDI.compute())
Here is my code 02: Using dask.dataframe.
def DDlNDIwrap(df, data_name='nir'):
dtag = target_hexgrid(df, hex_res=12)
slon, slat, data = df[['lon','lat',data_name]].values.T
tlon, tlat = dtag[['lon','lat']].values.T
tout = lNDI(list(zip(slon, slat)), data)(tlon, tlat)
dout = pd.DataFrame(np.vstack([tlon,tlat,tout]).T,
columns=['lon','lat',data_name]
)
return dd.from_pandas(dout, npartitions=1)
n_splits=96
#--- ds is the sentinel-2 satellite dataset. Reading from .til file.
gd_chunked_lNDI = [ delayed(DDlNDIwrap)(ds) for ds in dsrc.to_delayed()]
gd_lNDI = delayed(dd.concat)(gd_chunked_lNDI, axis=0)
gd = gd_lNDI.compute().compute()
I suspected that unknown patterns are coming on the edge/side-effect. What I meant is that those points could be around the edge of each partition so that there have not enough data points for interpolation. I found that in Dask manual I could possibly be able to use map_overlap, map_partitions, and map_blocks to solve my question. But I keep failed it. Could someone help me to solve this?
PS:
Following is what I tried by using map_overlap function.
def maplNDIwrap(df, data_name='nir'):
dtag = target_hexgrid(df, hex_res=12)
slon, slat, data = df[['lon','lat',data_name]].values.T
tlon, tlat = dtag[['lon','lat']].values.T
tout = lNDI(list(zip(slon, slat)), data)(tlon, tlat)
print(len(tlon), len(tout))
dout = pd.DataFrame(np.vstack([tlon,tlat,tout]).T,
columns=['lon','lat',data_name]
)
return dout
dtag = target_hexgrid(dsrc.compute(), hex_res=12)
gd_map_lNDI = dsrc.map_overlap(maplNDIwrap,1,1, meta=type(dsrc))
print(len(dtag)) #--> output: 353678
gd_map_lNDI.compute() #--> Not match expected size
Updates:
Here I defined few function to generate synthetic dataset.
def lonlat(lon_min,lon_max, lat_min, lat_max, res=5):
xps = round((lon_max - lon_min)*110*1e3/res)
yps = round((lat_max - lat_min)*110*1e3/res)
return np.meshgrid(np.linspace(lon_min,lon_max,xps),
np.linspace(lat_min,lat_max,yps)
)
def xy_based_map(x,y):
x = np.pi*x/180
y = np.pi*y/180
return np.log10((1 - x / 3. + x ** 2 + (2*x*y) ** 3) * np.exp(-x ** 2 - y ** 2))
Using the similar method I provided above with the result below. You can definitely see there are lines in the interpolation outputs.
lon_min, lon_max, lat_min, lat_max = -70.42,-70.40,-30.42,-30.40
lon05, lat05 = lonlat(lon_min, lon_max, lat_min, lat_max,res=5)
z05 = xy_based_map(lon05,lat05)
df05= pd.DataFrame(np.vstack((lon05.ravel(), lat05.ravel(), z05.ravel())).T, columns=['lon','lat','z'])
df05= dd.from_pandas(df05, npartitions=n_splits)
lon30, lat30 = lonlat(lon_min, lon_max, lat_min, lat_max,res=30)
z30 = xy_based_map(lon30,lat30)
df30= pd.DataFrame(np.vstack((lon30.ravel(), lat30.ravel(), z30.ravel())).T, columns=['lon','lat','z'])
df30= dd.from_pandas(df30, npartitions=n_splits)
```.
```python
tlon, tlat = df05[['lon','lat']].values.T
slon, slat, data = df10.values.T
gd_chunked_lNDI = [delayed(lNDIwrap)(x1, y1, newarr, xx, yy) for x1, y1, newarr, xx, yy in \
zip(slon.to_delayed().flatten(),
slat.to_delayed().flatten(),
data.to_delayed().flatten(),
tlon.to_delayed().flatten(),
tlat.to_delayed().flatten())]
gd_chunked_cNDI = [delayed(cNDIwrap)(x1, y1, newarr, xx, yy) for x1, y1, newarr, xx, yy in \
zip(slon.to_delayed().flatten(),
slat.to_delayed().flatten(),
data.to_delayed().flatten(),
tlon.to_delayed().flatten(),
tlat.to_delayed().flatten())]
gd_chunked_rNDI = [delayed(rNDIwrap)(x1, y1, newarr, xx, yy) for x1, y1, newarr, xx, yy in \
zip(slon.to_delayed().flatten(),
slat.to_delayed().flatten(),
data.to_delayed().flatten(),
tlon.to_delayed().flatten(),
tlat.to_delayed().flatten())]
gd_lNDI = delayed(da.concatenate)(gd_chunked_lNDI, axis=0)
gd_cNDI = delayed(da.concatenate)(gd_chunked_cNDI, axis=0)
gd_rNDI = delayed(da.concatenate)(gd_chunked_rNDI, axis=0)
results_lNDI_10m = np.array(gd_lNDI.compute())
results_cNDI_10m = np.array(gd_cNDI.compute())
results_rNDI_10m = np.array(gd_rNDI.compute())
#--- No parallel computing
a,b,c,d,e = slon.compute(),slat.compute(),data.compute(),tlon.compute(),tlat.compute()
straight_lNDI_10m = lNDIwrap(a,b,c,d,e)
###--- 30m --> 5m
tlon, tlat = df05[['lon','lat']].values.T
slon, slat, data = df30.values.T
gd_chunked_lNDI = [delayed(lNDIwrap)(x1, y1, newarr, xx, yy) for x1, y1, newarr, xx, yy in \
zip(slon.to_delayed().flatten(),
slat.to_delayed().flatten(),
data.to_delayed().flatten(),
tlon.to_delayed().flatten(),
tlat.to_delayed().flatten())]
gd_chunked_cNDI = [delayed(cNDIwrap)(x1, y1, newarr, xx, yy) for x1, y1, newarr, xx, yy in \
zip(slon.to_delayed().flatten(),
slat.to_delayed().flatten(),
data.to_delayed().flatten(),
tlon.to_delayed().flatten(),
tlat.to_delayed().flatten())]
gd_chunked_rNDI = [delayed(rNDIwrap)(x1, y1, newarr, xx, yy) for x1, y1, newarr, xx, yy in \
zip(slon.to_delayed().flatten(),
slat.to_delayed().flatten(),
data.to_delayed().flatten(),
tlon.to_delayed().flatten(),
tlat.to_delayed().flatten())]
gd_lNDI = delayed(da.concatenate)(gd_chunked_lNDI, axis=0)
gd_cNDI = delayed(da.concatenate)(gd_chunked_cNDI, axis=0)
gd_rNDI = delayed(da.concatenate)(gd_chunked_rNDI, axis=0)
results_lNDI_30m = np.array(gd_lNDI.compute())
results_cNDI_30m = np.array(gd_cNDI.compute())
results_rNDI_30m = np.array(gd_rNDI.compute())
###--- No Parallel for 30m --> 5m
a,b,c,d,e = slon.compute(),slat.compute(),data.compute(),tlon.compute(),tlat.compute()
straight_lNDI_30m = lNDIwrap(a,b,c,d,e)
###--- For plots.
dout= pd.DataFrame(np.vstack((tlon.compute(),tlat.compute(),df05.z.values.compute(),
results_lNDI_10m,results_cNDI_10m, results_rNDI_10m,straight_lNDI_10m,
results_lNDI_30m,results_cNDI_30m, results_rNDI_30m,straight_lNDI_30m,
)).T, columns=['lon','lat','orig','lNDI10','cNDI10','rNDI10','stgh10','lNDI30','cNDI30','rNDI30','stgh30'])