I have a dataframe of repetaed measures in WIDE format, with an ID variable (pid) some non time-varyant variables (i.e. age and sex) and mostly time-varyant variables, all named similarly with the following scheme: spec_var_0 spec_var_1 and spec_var_2 (we have 3 timewaves). Here it is an example:
structure(list(pid = structure(1:6, .Label = c("1", "2", "3",
"4", "5", "6", "7", "8", "9", "10", "11", "12", "13", "14", "15",
"16", "17", "18", "19", "20"), class = "factor"), dem_age = c(50,
73, 55, 66, 65, 70), dem_sex = structure(c(1L, 2L, 1L, 2L, 2L,
2L), .Label = c("Female", "Male"), class = "factor"), hx_steatosis = structure(c(1L,
1L, 1L, 1L, 1L, 1L), .Label = c("No", "Yes"), class = "factor"),
bio_weight_0 = c(89.4, 73.9, 78.7, 86.2, 132, 90.5), bio_weight_1 = c(94.9,
66, 75.2, 82, 119, 81.6315789473684), bio_weight_2 = c(92,
56, 71, 81, 118, 86.5), lab_glyc_0 = c(5.52545293913178,
4.88280192258637, 4.79579054559674, 4.94875989037817, 5.17614973257383,
5.0998664278242), lab_glyc_1 = c(5.08140436498446, 5.01063529409626,
5.01727983681492, 4.77912349311153, 4.95582705760126, 5.03695260241363
), lab_glyc_2 = c(4.39444915467244, 5.06890420222023, 4.52178857704904,
4.62497281328427, 5.34710753071747, 4.95582705760126)), row.names = c(NA,
6L), class = "data.frame")
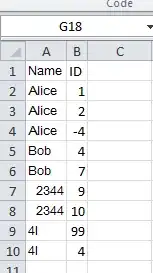
Now I would like to convert to pivot_longer so to have the following format (sorry for the image, I do not know how to build such a table in R)
Searching Stack and other forums it seems that the issue could be fixed using refular expressions,but I have no idea of how to use them (in general and in this context specifically).
Any help would be (as usual) greatly appreciated