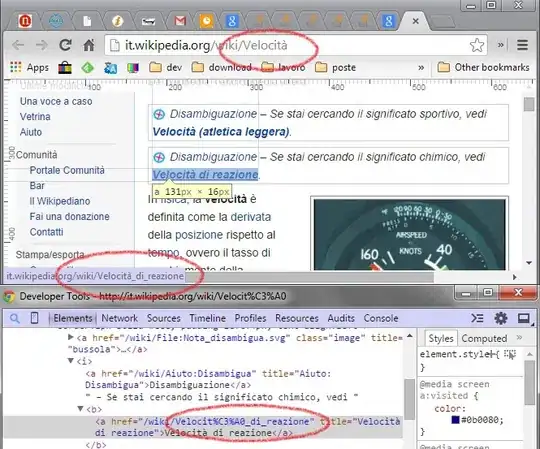
I'm working with the wisconsin breast cancer dataset found here. Feature engineering is important in machine learning so a teacher of mine recommended the MeanEncoder part of a library found here. The dataframe looks like the following:
I did specifically change the diagnosis feature/column to category because one of the errors said that might of been the issue but apparently not as it's not solved.
I want to mean encode the target feature/column using MeanEncode found in the library linked above. Here's my function to attempt to do so:
def MeanEncoding(self):
# Get the columns besides the target variable at the front, which is diagnosis, as recommended by teacher.
cols = self.m_df.iloc[:, 1:].columns.to_list()
# Save specifically the target variable too.
target = self.m_df.iloc[:, 0]
# Now get the object ready.
encoder = MeanEncoder(variables=cols)
print('---Fitting---')
encoder.fit(self.m_df.drop('diagnosis', axis=1), target)
In this code:
- m_df - just the dataframe hence the "df"
- I drop the diagnosis column/feature in the first argument of encoder.fit, since it's provided in the 2nd argument of the same function. But it means nothing. Because I still get the error: "TypeError: Some of the variables are not categorical. Please cast them as object or category before calling this transformer"
Now with #2, I'm thinking, "No way, I have to transform the numeric features which are 'radius_mean', 'texture_mean', etc into category or object? That makes 0 sense". But I google this error of course and it brings me to this SO thread. This individual is having similar concerns like me except with a different function. The suggestion for him was "Just change the dtype of grade column to object before using imputer", so I change the types as well to object with the following code:
for i in range(1, len(self.m_df.columns)):
columnName = self.m_df.columns[i]
self.m_df[columnName] = self.m_df[columnName].astype('object')
Doesn't make sense to me because it's converting the types of genuine numeric columns/features. I get this error which is KIND of expected:
pandas.core.base.DataError: No numeric types to aggregate
Now I'm thinking it just wants a few numeric types, so I slightly alter the code:
for i in range(1, len(self.m_df.columns) - 2):
columnName = self.m_df.columns[i]
self.m_df[columnName] = self.m_df[columnName].astype('object')
Which literally just leaves the last 2 columns as float64 types and therefore all others are type object (besides the diagnosis column which is category but I doubt that matters). Now some numeric types ARE present. Yet I still get the error again
TypeError: Some of the variables are not categorical. Please cast
them as object or category before calling this transformer
I am clearly missing something but not sure what. No matter how I alter the types to satisfy the function, it's wrong.