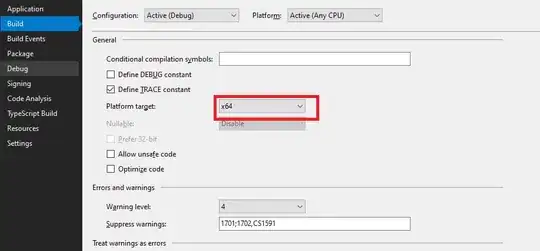
Does this example help?
library(caret)
library(mlr)
library(MASS)
Boston1 <- Boston[,9:14]
datasplit <- createDataPartition(y = Boston1$medv, times = 1,
p = 0.75, list = FALSE)
Boston1_train <- Boston1[datasplit,]
Boston1_test <- Boston1[-datasplit,]
rtask <- makeRegrTask(id = "bh", data = Boston1_train, target = "medv")
rmod <- train(makeLearner("regr.lm"), rtask)
rmod1 <- train(makeLearner("regr.fnn"), rtask)
rmod2 <- train(makeLearner("regr.randomForest"), rtask)
plotLearnerPrediction("regr.lm", features = "lstat", task = rtask)
plotLearnerPrediction("regr.fnn", features = "lstat", task = rtask)
plotLearnerPrediction("regr.randomForest", features = "lstat", task = rtask)
# what other learners could be used?
# lrns = listLearners()
rmod3 <- train(makeLearner("regr.cforest"), rtask)
rmod4 <- train(makeLearner("regr.ksvm"), rtask)
rmod5 <- train(makeLearner("regr.glmboost"), rtask)
plotLearnerPrediction("regr.cforest", features = "lstat", task = rtask)
plotLearnerPrediction("regr.ksvm", features = "lstat", task = rtask)
plotLearnerPrediction("regr.glmboost", features = "lstat", task = rtask)

pred_mod = predict(rmod, newdata = Boston1_test)
pred_mod1 = predict(rmod1, newdata = Boston1_test)
pred_mod2 = predict(rmod2, newdata = Boston1_test)
pred_mod3 = predict(rmod3, newdata = Boston1_test)
pred_mod4 = predict(rmod4, newdata = Boston1_test)
pred_mod5 = predict(rmod5, newdata = Boston1_test)
predictions <- data.frame(truth = pred_mod$data$truth,
regr.lm = pred_mod$data$response,
regr.fnn = pred_mod1$data$response,
regr.randomForest = pred_mod2$data$response,
regr.cforest = pred_mod3$data$response,
regr.ksvm = pred_mod4$data$response,
regr.glmboost = pred_mod5$data$response)
library(tidyverse)
predictions %>%
pivot_longer(-truth, values_to = "predicted_value") %>%
ggplot(aes(x = truth, y = predicted_value, color = name)) +
geom_smooth() +
geom_abline(slope = 1, intercept = c(0, 0))