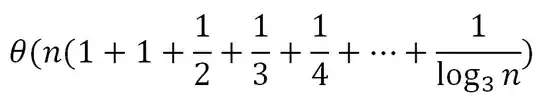I have a dataframe with 40 rows and ~40000 columns. The 40 rows are split into group "A" and group "B" (20 each). For each column, I would like to apply a statistical test (wilcox.test()) comparing the two groups. I started using a for loop to run through the 40000 columns but it was very slow.
Minimal Reproducible Example (MRE):
library(tidyverse)
set.seed(123)
metrics <- paste("metric_", 1:40000, sep = "")
patient_IDs <- paste("patientID_", 1:40, sep = "")
m <- matrix(sample(1:20, 1600000, replace = TRUE), ncol = 40000, nrow = 40,
dimnames=list(patient_IDs, metrics))
test_data <- as.data.frame(m)
test_data$group <- c(rep("A", 20), rep("B", 20))
# Collate list of metrics to analyse ("check") for significance
list_to_check <- colnames(test_data)[1:40000]
Original 'loop' method (this is what I want to vectorise):
# Create a variable to store the results
results_A_vs_B <- c()
# Loop through the "list_to_check" and,
# for each 'value', compare group A with group B
# and load the results into the "results_A_vs_B" variable
for (i in list_to_check) {
outcome <- wilcox.test(test_data[test_data$group == "A", ][[i]],
test_data[test_data$group == "B", ][[i]],
exact = FALSE)
if (!is.nan(outcome$p.value) && outcome$p.value <= 0.05) {
results_A_vs_B[i] <- paste(outcome$p.value, sep = "\t")
}
}
# Format the results into a dataframe
summarised_results_A_vs_B <- as.data.frame(results_A_vs_B) %>%
rownames_to_column(var = "A vs B") %>%
rename("Wilcox Test P-value" = "results_A_vs_B")
Benchmarking the answers so far:
# Ronak Shah's "Map" approach
Map_func <- function(dataset, list_to_check) {
tmp <- split(dataset[list_to_check], dataset$group)
stack(Map(function(x, y) wilcox.test(x, y, exact = FALSE)$p.value, tmp[[1]], tmp[[2]]))
}
# @Onyambu's data.table method
dt_func <- function(dataset, list_to_check) {
melt(setDT(dataset), measure.vars = list_to_check)[, dcast(.SD, rowid(group) + variable ~ group)][, wilcox.test(A, B, exact = FALSE)$p.value, variable]
}
# @Park's dplyr method (with some minor tweaks)
dplyr_func <- function(dataset, list_to_check){
dataset %>%
summarise(across(all_of(list_to_check),
~ wilcox.test(.x ~ group, exact = FALSE)$p.value)) %>%
pivot_longer(cols = everything(),
names_to = "Metrics",
values_to = "Wilcox Test P-value")
}
library(microbenchmark)
res_map <- microbenchmark(Map_func(test_data, list_to_check), times = 10)
res_dplyr <- microbenchmark(dplyr_func(test_data, list_to_check), times = 2)
library(data.table)
res_dt <- microbenchmark(dt_func(test_data, list_to_check), times = 10)
autoplot(rbind(res_map, res_dt, res_dplyr))
# Excluding dplyr
autoplot(rbind(res_map, res_dt))
--
Running the code on a server took a couple of seconds longer but the difference between Map and data.table was more pronounced (laptop = 4 cores, server = 8 cores):
autoplot(rbind(res_map, res_dt))