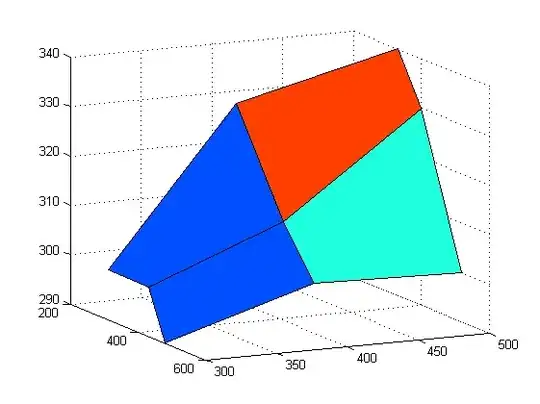
I have point data off the coast of Oahu. Someone else used those same data to create a large polygon. I believe he first created a heatmap using a quartic (biweight) kernel with a radius of 1 km around each point and perhaps a 1 km-square pixel size. He cited Silverman (1986, p. 76, equation 4.5, which I believe refers to the book “Density Estimation for Statistics and Data Analysis”). I believe he converted his heatmap into his polygon. I am attempting to approximate his polygon with fake data using R and Windows 10. I can come close using the kde function in the ks package (see figure below). But that package only includes Gaussian kernels. Is it possible to create a similar polygon using a quartic kernel?
The other analysist actually created two version of the polygon. The border of one was labeled “> 1 per km density”; the border of the other was labeled “> 0.5 per km density”. I do not know whether he used R, QGIS, ArcGIS or something else. I was unable to create a single large polygon in QGIS and do not have ArcGIS.
Thank you for any suggestions on how to create a polygon similar to the one shown but using a quartic kernel instead of a Gaussian kernel. If I can provide additional information please let me know.
Here is a link to my fake data in CSV and QGIS format: enter link description here (EDIT: Hopefully anyone can access the fake data now. I could before but I guess others could not.)
1. fake_points_oahu.csv
a. raw data
2. fake_points_oahu_utm (.shp, .dbf, .prj, .shx)
a. vector point layer
3. fake_points_oahu_June11_2021.png
a. the figure shown above
Here is my R code:
setwd('C:/Users/mark_/Documents/ctmm/density_in_R/density_files_for_StackOverflow/')
library(sf) # to read shapefile
library(ks) # to use kde function
my.data <- read.csv("fake_points_oahu.csv", header = TRUE, stringsAsFactors = FALSE, na.strings = "NA")
head(my.data)
# Import shapefile
st_layers("fake_points_oahu_utm.shp")
points_utm <- st_read(dsn = "fake_points_oahu_utm.shp", layer = 'fake_points_oahu_utm')
st_crs(points_utm)
plot(points_utm)
my.matrix <- as.matrix(my.data[,2:3])
head(my.matrix)
# This uses the Guassian kernel
my_gps_hpi <- Hpi(x = my.matrix, pilot = "samse", pre = "scale")
my.fhat <- kde(x = my.matrix, compute.cont = TRUE, h = my_gps_hpi,
xmin = c(min(my.data$longitude), min(my.data$latitude)),
xmax = c(max(my.data$longitude), max(my.data$latitude)),
bgridsize = c(500, 500))
my.contours <- c(96.5)
contourLevels(my.fhat, cont = my.contours)
contourSizes(my.fhat, cont = my.contours, approx = TRUE)
plot(my.data$longitude, my.data$latitude)
plot(my.fhat, lwd = 3, display = "filled.contour", cont = my.contours, add = TRUE)
png(file="fake_points_oahu_June11_2021.png")
plot(my.data$longitude, my.data$latitude)
plot(my.fhat, lwd = 3, display = "filled.contour", cont = my.contours, add = TRUE)
dev.off()