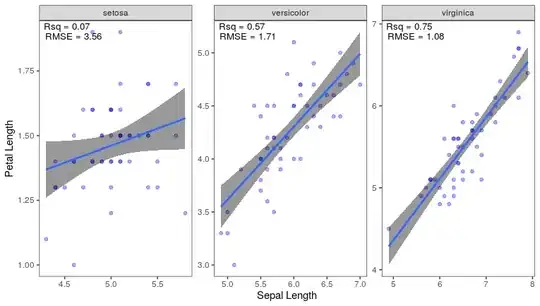
I am trying to automatically position R2 and RMSE in facetted ggplot. This answer has helped me to put the R2 and RMSE in facetted ggplot manually using the following code
library(caret)
library(tidyverse)
summ <- iris %>%
group_by(Species) %>%
summarise(Rsq = R2(Sepal.Length, Petal.Length),
RMSE = RMSE(Sepal.Length, Petal.Length)) %>%
mutate_if(is.numeric, round, digits=2)
p <- ggplot(data=iris, aes(x = Sepal.Length, y = Petal.Length)) +
geom_point(color="blue",alpha = 1/3) +
facet_wrap(Species ~ ., scales="free") +
geom_smooth(method=lm, fill="black", formula = y ~ x) +
xlab("Sepal Length") +
ylab("Petal Length") + theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
# Here we create our annotations data frame.
df.annotations <- data.frame()
# Rsq
df.annotations <- rbind(df.annotations,
cbind(as.character(summ$Species),
paste("Rsq", summ$Rsq,
sep = " = ")))
# RMSE
df.annotations <- rbind(df.annotations,
cbind(as.character(summ$Species),
paste("RMSE", summ$RMSE,
sep = " = ")))
# This here is important, especially naming the first column
# Species
colnames(df.annotations) <- c("Species", "label")
df.annotations$x <- rep.int(c(4.5, 5.5, 5.5), times = 2)
df.annotations$y <- c(1.75, 5.0, 6.8,
1.7, 4.9, 6.7)
p + geom_text(data = df.annotations,
mapping = aes(x = x, y = y, label = label))
As you can see from df.annotations, I am providing the positions manually. Now how can I automatically position R2 and RMSE in ggplot2?