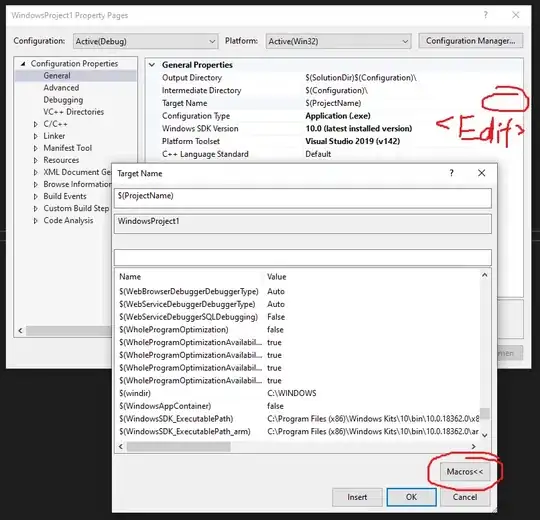
How to account for paired observations in statistical tests other than t-test? Below I discuss two examples in which I try to do so with a mixed-effect approach and fail.
Example 1: how to reproduce t.test(..., paired=T) in lm()?
# simulate data
set.seed(66)
x1 <- rnorm(n=100, mean=30, sd=6)
x2 <- rnorm(n=100, mean=60, sd=6)
# arrange the data in a dataset
dd <- data.frame(ID=rep(paste("ID", seq(1,100, by=1), sep="_"),2),
response=c(x1,x2),
group=c(rep("A", 100), rep("B", 100))
)
t.test(x1,x2, paired=F)
summary(lm(response~group, data=dd)) # same outcome
If the observations are paired one can account for it with t.test() but how to do so in lm() (if at all possible)? I tried to use a mixed-effect model approach, but:
summary(lmerTest::lmer(response~group + (1+group|ID), data=dd))
Gives an error:
Error: number of observations (=200) <= number of random effects (=200) for term (1 + group | ID);
the random-effects parameters and the residual variance (or scale parameter) are probably unidentifiable
While:
summary(lmerTest::lmer(response~group + (1|ID), data=dd))
Runs but the fixed effect parameter estimates and associated Std. Errors and t values are identical to those produced by lm().
Example 2: linear regression with paired observations
Let's imagine that the observations in the dataset I created were from subjects measured 30 days apart - namely, each of the 100 subjects were measured on day 0, then again on day 30 - and we wanted to estimate the rate of change over time:
dd$time=c(rep(0,100), rep(30, 100)) # add "time" variable to dd
Data look like this (linear regression in black, paired data linked by red lines):
lm1 <- lm(response~time, data=dd)
lm1 does not account for the paired nature of the observations.
I thought of running a mixed-effect model that allowed for each pair of data to differ in intercept and slope, but R protests again that I am trying to estimate too many parameters:
lmerTest::lmer(response ~ time + (time | ID), data=dd)
# Error: number of observations (=200) <= number of random effects (=200) for term (time | ID);
# the random-effects parameters and the residual variance (or scale parameter) are probably unidentifiable
A simpler model that allows data pairs to differ in intercept but not in slope, namely:
lmer(response ~ time + (1 | ID), data=dd)
Complains that:
boundary (singular) fit: see ?isSingular
But runs and produces fixed effect estimates that are identical to those produced by lm().
[UPDATE]
@Limey reminded me that a paired t-test is nothing but a t-test assessing whether the pairwise differences between the two groups differ from zero. Such pairwise difference could be used to perform any paired statistical test beside a t test. To verify this I created three different "Response" variables that are a combination of x1 and x2 ordered in different ways (respectively: original random order; x1 in increasing and x2 in decreasing order; both in increasing order).
dd$response2 <- c(sort(x1, decreasing = FALSE), sort(x2, decreasing = T))
dd$response3 <- c(sort(x1, decreasing = FALSE), sort(x2, decreasing = F))
I calculated the corresponding differences:
dd$diff1 <- c((dd$response[1:100]-dd$response[1:100]),
(dd$response[101:200]-dd$response[1:100]))
dd$diff2 <- c((dd$response2[1:100]-dd$response2[1:100]),
(dd$response2[101:200]-dd$response2[1:100]))
dd$diff3 <- c((dd$response3[1:100]-dd$response3[1:100]),
(dd$response3[101:200]-dd$response3[1:100]))
 And used them to perform linear models:
And used them to perform linear models:
lm2.diff1 <- lm(diff1~time, data=dd)
lm2.diff2 <- lm(diff2 ~time, data=dd)
lm2.diff3 <- lm(diff3 ~time, data=dd)
I expected them to differ in their slope estimates, but they were all the same:
summary(lm2.diff1)$coeff[2] # 0.9993754
summary(lm2.diff2)$coeff[2] # 0.9993754
summary(lm2.diff3)$coeff[2] # 0.9993754
Their slope estimate is the same estimated from the corresponding "unpaired" linear models (lm(response~time), lm(response2~time), and lm(response3~time)). What am I missing?