Everyone I made a machine learning project using binary patterns to detect plant diseases using haralick textures from images, I trained it with 5 different set of data and it predicting in 60% accuracy. Now i got a situation that to print 3 probable diseases on one image. Example i uploaded one image and predicted it got ‘Mites’ and also want to check is there any other 3 probable diseases in image of plant.
how to achieve the 3 probable in python using local binary patterns?
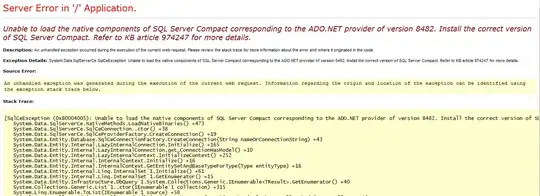
Full code am trying
import cv2
import numpy as np
import os
import glob
import mahotas as mt
from sklearn.svm import LinearSVC
from sklearn.metrics import mean_squared_error
import joblib
# function to extract haralick textures from an image
def extract_features(image):
# calculate haralick texture features for 4 types of adjacency
textures = mt.features.haralick(image)
# take the mean of it and return it
ht_mean = textures.mean(axis=0)
return ht_mean
def ResizeWithAspectRatio(image, width=None, height=None, inter=cv2.INTER_AREA):
dim = None
(h, w) = image.shape[:2]
if width is None and height is None:
return image
if width is None:
r = height / float(h)
dim = (int(w * r), height)
else:
r = width / float(w)
dim = (width, int(h * r))
return cv2.resize(image, dim, interpolation=inter)
# load the training dataset
train_path = "D:/ai training/aphids/Anothertest"
train_names = os.listdir(train_path)
# empty list to hold feature vectors and train labels
train_features = []
train_labels = []
# loop over the training dataset
print ("[STATUS] Started extracting haralick textures..")
for train_name in train_names:
cur_path = train_path + "/" + train_name
cur_label = train_name
i = 1
for file in glob.glob(cur_path + "/*.jpg"):
print ("Processing Image - {} in {}".format(i, cur_label))
# read the training image
image = cv2.imread(file)
resize = ResizeWithAspectRatio(image, width=1250, height=1000) # Resize by width OR
# convert the image to grayscale
gray = cv2.cvtColor(resize, cv2.COLOR_BGR2GRAY)
# extract haralick texture from the image
features = extract_features(gray)
# append the feature vector and label
train_features.append(features)
train_labels.append(cur_label)
# otherwise create the model, train the model and save the model
if os.path.exists("D:/ai training/aphids/joblib_model.sav"):
print("Loading Trained Model")
clf_svm = joblib.load("D:/ai training/aphids/Anothertest/joblib_model.sav")
else:
# have a look at the size of our feature vector and labels
print ("Training features: {}".format(np.array(train_features).shape))
print ("Training labels: {}".format(np.array(train_labels).shape))
# create the classifier
print ("[STATUS] Creating the classifier..")
clf_svm = LinearSVC(random_state=9, dual=False, max_iter=1000)
# fit the training data and labels
print ("[STATUS] Fitting data/label to model..")
clf_svm.fit(train_features, train_labels)
#savemodel
joblib_file = 'D:/ai training/aphids/joblib_model.sav'
joblib.dump(clf_svm, joblib_file)
#epoch
#clf_svm.fit(train_features, train_labels, epochs=10, validation_data=(X_test), y_test), batch_size=64)
#clf_svm.fit(train_features, train_labels, epochs=10, validation_data=(X_test, y_test), batch_size=64)
# loop over the test images
test_path = "D:/ai training/aphids/tata"
for file in glob.glob(test_path + "/*.jpg"):
# read the input image
image = cv2.imread(file)
resize = ResizeWithAspectRatio(image, width=1250, height=1000) # Resize by width OR
# convert to grayscale
gray = cv2.cvtColor(resize, cv2.COLOR_BGR2GRAY)
# extract haralick texture from the image
features = extract_features(gray)
# evaluate the model and predict label
prediction = clf_svm.predict(features.reshape(1, -1))[0]
clf_svm.fit(train_features, train_labels)
# show the label
cv2.putText(resize, prediction, (10,30), cv2.FONT_HERSHEY_SIMPLEX, 1.0, (0,0,255), 3)
print ("Prediction - {}".format(prediction))
print("Accuracy - ", clf_svm.score(train_features, train_labels))
# display the output image
cv2.namedWindow
cv2.imshow("Test_Image", resize)
cv2.waitKey(0)