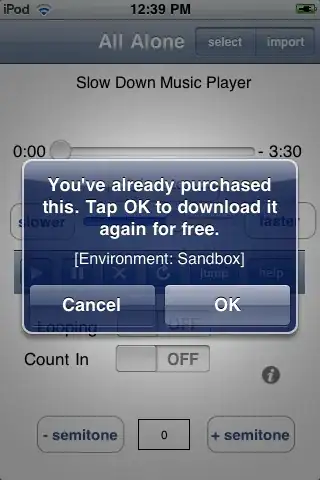
Please take a look at the image A below.  Some introduction to my question:
My goal is to obtain an accurate coordinate trace of the illustrated DNA molecule. The coordinates of the trace are represented by the blue dots in the image and are represented as a 2-d numpy array in Python, i.e.:
Some introduction to my question:
My goal is to obtain an accurate coordinate trace of the illustrated DNA molecule. The coordinates of the trace are represented by the blue dots in the image and are represented as a 2-d numpy array in Python, i.e.: trace: nd-array; shape (N, 2) where N is the number of trace points. The plots was done using plt.scatter(trace[:, 1], trace[:, 0]).
Now, please take a closer look at the following function:
def rel_coords_to_trace(trace, distance_limit=5.0):
"""
Finds the pixels in the image that are within the 'distance_limit' of the 'trace' points. For those pixels the
relative coordinates to the closest trace point is calculated.
Args:
trace ([N, 2] array): Initial trace of the DNA strand
distance_limit (float): Maximum distance a pixel can have from the trace to be taken into account
Returns:
pixels: Array with row/column coordinates of the pixels within the distance limit from the trace
trace_id: Int relating each pixel from the 'pixels' array to the point in the 'trace' it is closest to
relative_coords ([N, 2] array): Relative x and y distances of all pixels from the closest point of the trace
heights([N, ] array): Height of the image at the position of the pixel
"""
min_r, min_c = np.floor(trace.min(axis=0) - distance_limit).astype(int).clip(min=0)
max_r, max_c = np.ceil(trace.max(axis=0) + distance_limit).astype(int).clip(max=mol_filtered.shape)
pixels_pos = np.mgrid[min_r:max_r, min_c:max_c].reshape([2, -1]).T # all potential pixels
# kdTree finds the nearest neighbour between a specific pixel and all trace points
# Returns distances between pixels and nn and the id of the nn. Distances are inf if bigger than distance_limit
kdtree = cKDTree(trace)
distances, trace_id = kdtree.query(pixels_pos, k=1, distance_upper_bound=distance_limit)
pixels = pixels_pos[distances != np.inf]
trace_id = trace_id[distances != np.inf]
rel_coords = pixels - trace[trace_id]
return rel_coords, pixels, trace_id
Its execution is illustrated in Image B My question:
Now, when I have a sharp turn in my coordinate trace, I get comparatively many white arrows that point to a specific trace point from more or less one direction. My goal is to quantify how many more white arrows are pointing from one side of the trace (in a direction normal to the trace) compared to the other side of the trace. This quantification doesn't have to be exact, I just somehow want to add a respective weight into the mix.
My question:
Now, when I have a sharp turn in my coordinate trace, I get comparatively many white arrows that point to a specific trace point from more or less one direction. My goal is to quantify how many more white arrows are pointing from one side of the trace (in a direction normal to the trace) compared to the other side of the trace. This quantification doesn't have to be exact, I just somehow want to add a respective weight into the mix.
How can I achieve this quantification?