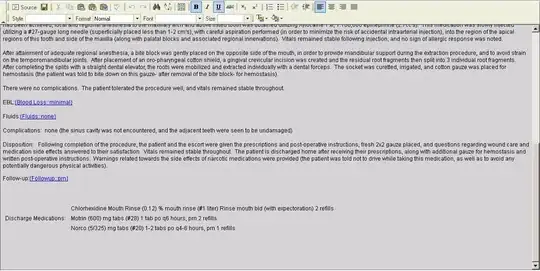
Is your data continuous or categorical? It seems to be categorical. It does not make a lot of sense to calculate distance between binary variables. Not all data is well-suited for clustering.
I don't have your actual data, but I'll show you how to do clustering correctly, and incorrectly, using the canonical MTCars sample data.
# import mtcars data from web, and do some clustering on the data set
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.cluster import MiniBatchKMeans
# Import CSV mtcars
data = pd.read_csv('https://gist.githubusercontent.com/ZeccaLehn/4e06d2575eb9589dbe8c365d61cb056c/raw/64f1660f38ef523b2a1a13be77b002b98665cdfe/mtcars.csv')
# Edit element of column header
data.rename(columns={'Unnamed: 0':'brand'}, inplace=True)
X1= data.iloc[:,1:12]
Y1= data.iloc[:,-1]
#lets try to plot Decision tree to find the feature importance
from sklearn.tree import DecisionTreeClassifier
tree= DecisionTreeClassifier(criterion='entropy', random_state=1)
tree.fit(X1, Y1)
imp= pd.DataFrame(index=X1.columns, data=tree.feature_importances_, columns=['Imp'] )
imp.sort_values(by='Imp', ascending=False)
sns.barplot(x=imp.index.tolist(), y=imp.values.ravel(), palette='coolwarm')
X=data[['cyl','drat']]
Y=data['carb']
#lets try to create segments using K means clustering
from sklearn.cluster import KMeans
#using elbow method to find no of clusters
wcss=[]
for i in range(1,7):
kmeans= KMeans(n_clusters=i, init='k-means++', random_state=1)
kmeans.fit(X)
wcss.append(kmeans.inertia_)
plt.plot(range(1,7), wcss, linestyle='--', marker='o', label='WCSS value')
plt.title('WCSS value- Elbow method')
plt.xlabel('no of clusters- K value')
plt.ylabel('Wcss value')
plt.legend()
plt.show()
kmeans.predict(X)
#Cluster Center
kmeans = MiniBatchKMeans(n_clusters = 5)
kmeans.fit(X)
centroids = kmeans.cluster_centers_
labels = kmeans.labels_
print(centroids)
print(labels)
colors = ["green", "red", "blue", "yellow", "orange"]
plt.scatter(X.iloc[:,0], X.iloc[:,1], c=np.array(colors)[labels], s = 10, alpha=.1)
plt.scatter(centroids[:, 0], centroids[:, 1], marker = "x", s=150, linewidths = 5, zorder = 10, c=colors)
plt.show()
...now, I am just changing two features (two independent variables)...and re-running the same experiment...
X=data[['wt','qsec']]
Y=data['carb']
#lets try to create segments using K means clustering
from sklearn.cluster import KMeans
#using elbow method to find no of clusters
wcss=[]
for i in range(1,7):
kmeans= KMeans(n_clusters=i, init='k-means++', random_state=1)
kmeans.fit(X)
wcss.append(kmeans.inertia_)
plt.plot(range(1,7), wcss, linestyle='--', marker='o', label='WCSS value')
plt.title('WCSS value- Elbow method')
plt.xlabel('no of clusters- K value')
plt.ylabel('Wcss value')
plt.legend()
plt.show()
kmeans.predict(X)
#Cluster Center
kmeans = MiniBatchKMeans(n_clusters = 5)
kmeans.fit(X)
centroids = kmeans.cluster_centers_
labels = kmeans.labels_
print(centroids)
print(labels)
colors = ["green", "red", "blue", "yellow", "orange"]
plt.scatter(X.iloc[:,0], X.iloc[:,1], c=np.array(colors)[labels], s = 10, alpha=.1)
plt.scatter(centroids[:, 0], centroids[:, 1], marker = "x", s=150, linewidths = 5, zorder = 10, c=colors)
plt.show()
As you can see, the choice of features that you use for clustering makes a huge difference in the outcome (obviously). The first example looks somewhat like your results and the second example looks like a more useful/interesting clustering experiment.