I am attempting to generate map overlay images that would assist in identifying hot-spots, that is areas on the map that have high density of data points. None of the approaches that I've tried are fast enough for my needs. Note: I forgot to mention that the algorithm should work well under both low and high zoom scenarios (or low and high data point density).
I looked through numpy, pyplot and scipy libraries, and the closest I could find was numpy.histogram2d. As you can see in the image below, the histogram2d output is rather crude. (Each image includes points overlaying the heatmap for better understanding)
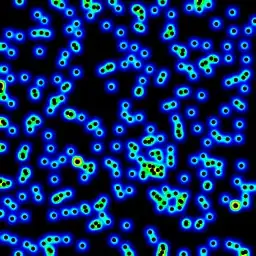
 My second attempt was to iterate over all the data points, and then calculate the hot-spot value as a function of distance. This produced a better looking image, however it is too slow to use in my application. Since it's O(n), it works ok with 100 points, but blows out when I use my actual dataset of 30000 points.
My second attempt was to iterate over all the data points, and then calculate the hot-spot value as a function of distance. This produced a better looking image, however it is too slow to use in my application. Since it's O(n), it works ok with 100 points, but blows out when I use my actual dataset of 30000 points.
My final attempt was to store the data in an KDTree, and use the nearest 5 points to calculate the hot-spot value. This algorithm is O(1), so much faster with large dataset. It's still not fast enough, it takes about 20 seconds to generate a 256x256 bitmap, and I would like this to happen in around 1 second time.
Edit
The boxsum smoothing solution provided by 6502 works well at all zoom levels and is much faster than my original methods.
The gaussian filter solution suggested by Luke and Neil G is the fastest.
You can see all four approaches below, using 1000 data points in total, at 3x zoom there are around 60 points visible.

Complete code that generates my original 3 attempts, the boxsum smoothing solution provided by 6502 and gaussian filter suggested by Luke (improved to handle edges better and allow zooming in) is here:
import matplotlib
import numpy as np
from matplotlib.mlab import griddata
import matplotlib.cm as cm
import matplotlib.pyplot as plt
import math
from scipy.spatial import KDTree
import time
import scipy.ndimage as ndi
def grid_density_kdtree(xl, yl, xi, yi, dfactor):
zz = np.empty([len(xi),len(yi)], dtype=np.uint8)
zipped = zip(xl, yl)
kdtree = KDTree(zipped)
for xci in range(0, len(xi)):
xc = xi[xci]
for yci in range(0, len(yi)):
yc = yi[yci]
density = 0.
retvalset = kdtree.query((xc,yc), k=5)
for dist in retvalset[0]:
density = density + math.exp(-dfactor * pow(dist, 2)) / 5
zz[yci][xci] = min(density, 1.0) * 255
return zz
def grid_density(xl, yl, xi, yi):
ximin, ximax = min(xi), max(xi)
yimin, yimax = min(yi), max(yi)
xxi,yyi = np.meshgrid(xi,yi)
#zz = np.empty_like(xxi)
zz = np.empty([len(xi),len(yi)])
for xci in range(0, len(xi)):
xc = xi[xci]
for yci in range(0, len(yi)):
yc = yi[yci]
density = 0.
for i in range(0,len(xl)):
xd = math.fabs(xl[i] - xc)
yd = math.fabs(yl[i] - yc)
if xd < 1 and yd < 1:
dist = math.sqrt(math.pow(xd, 2) + math.pow(yd, 2))
density = density + math.exp(-5.0 * pow(dist, 2))
zz[yci][xci] = density
return zz
def boxsum(img, w, h, r):
st = [0] * (w+1) * (h+1)
for x in xrange(w):
st[x+1] = st[x] + img[x]
for y in xrange(h):
st[(y+1)*(w+1)] = st[y*(w+1)] + img[y*w]
for x in xrange(w):
st[(y+1)*(w+1)+(x+1)] = st[(y+1)*(w+1)+x] + st[y*(w+1)+(x+1)] - st[y*(w+1)+x] + img[y*w+x]
for y in xrange(h):
y0 = max(0, y - r)
y1 = min(h, y + r + 1)
for x in xrange(w):
x0 = max(0, x - r)
x1 = min(w, x + r + 1)
img[y*w+x] = st[y0*(w+1)+x0] + st[y1*(w+1)+x1] - st[y1*(w+1)+x0] - st[y0*(w+1)+x1]
def grid_density_boxsum(x0, y0, x1, y1, w, h, data):
kx = (w - 1) / (x1 - x0)
ky = (h - 1) / (y1 - y0)
r = 15
border = r * 2
imgw = (w + 2 * border)
imgh = (h + 2 * border)
img = [0] * (imgw * imgh)
for x, y in data:
ix = int((x - x0) * kx) + border
iy = int((y - y0) * ky) + border
if 0 <= ix < imgw and 0 <= iy < imgh:
img[iy * imgw + ix] += 1
for p in xrange(4):
boxsum(img, imgw, imgh, r)
a = np.array(img).reshape(imgh,imgw)
b = a[border:(border+h),border:(border+w)]
return b
def grid_density_gaussian_filter(x0, y0, x1, y1, w, h, data):
kx = (w - 1) / (x1 - x0)
ky = (h - 1) / (y1 - y0)
r = 20
border = r
imgw = (w + 2 * border)
imgh = (h + 2 * border)
img = np.zeros((imgh,imgw))
for x, y in data:
ix = int((x - x0) * kx) + border
iy = int((y - y0) * ky) + border
if 0 <= ix < imgw and 0 <= iy < imgh:
img[iy][ix] += 1
return ndi.gaussian_filter(img, (r,r)) ## gaussian convolution
def generate_graph():
n = 1000
# data points range
data_ymin = -2.
data_ymax = 2.
data_xmin = -2.
data_xmax = 2.
# view area range
view_ymin = -.5
view_ymax = .5
view_xmin = -.5
view_xmax = .5
# generate data
xl = np.random.uniform(data_xmin, data_xmax, n)
yl = np.random.uniform(data_ymin, data_ymax, n)
zl = np.random.uniform(0, 1, n)
# get visible data points
xlvis = []
ylvis = []
for i in range(0,len(xl)):
if view_xmin < xl[i] < view_xmax and view_ymin < yl[i] < view_ymax:
xlvis.append(xl[i])
ylvis.append(yl[i])
fig = plt.figure()
# plot histogram
plt1 = fig.add_subplot(221)
plt1.set_axis_off()
t0 = time.clock()
zd, xe, ye = np.histogram2d(yl, xl, bins=10, range=[[view_ymin, view_ymax],[view_xmin, view_xmax]], normed=True)
plt.title('numpy.histogram2d - '+str(time.clock()-t0)+"sec")
plt.imshow(zd, origin='lower', extent=[view_xmin, view_xmax, view_ymin, view_ymax])
plt.scatter(xlvis, ylvis)
# plot density calculated with kdtree
plt2 = fig.add_subplot(222)
plt2.set_axis_off()
xi = np.linspace(view_xmin, view_xmax, 256)
yi = np.linspace(view_ymin, view_ymax, 256)
t0 = time.clock()
zd = grid_density_kdtree(xl, yl, xi, yi, 70)
plt.title('function of 5 nearest using kdtree\n'+str(time.clock()-t0)+"sec")
cmap=cm.jet
A = (cmap(zd/256.0)*255).astype(np.uint8)
#A[:,:,3] = zd
plt.imshow(A , origin='lower', extent=[view_xmin, view_xmax, view_ymin, view_ymax])
plt.scatter(xlvis, ylvis)
# gaussian filter
plt3 = fig.add_subplot(223)
plt3.set_axis_off()
t0 = time.clock()
zd = grid_density_gaussian_filter(view_xmin, view_ymin, view_xmax, view_ymax, 256, 256, zip(xl, yl))
plt.title('ndi.gaussian_filter - '+str(time.clock()-t0)+"sec")
plt.imshow(zd , origin='lower', extent=[view_xmin, view_xmax, view_ymin, view_ymax])
plt.scatter(xlvis, ylvis)
# boxsum smoothing
plt3 = fig.add_subplot(224)
plt3.set_axis_off()
t0 = time.clock()
zd = grid_density_boxsum(view_xmin, view_ymin, view_xmax, view_ymax, 256, 256, zip(xl, yl))
plt.title('boxsum smoothing - '+str(time.clock()-t0)+"sec")
plt.imshow(zd, origin='lower', extent=[view_xmin, view_xmax, view_ymin, view_ymax])
plt.scatter(xlvis, ylvis)
if __name__=='__main__':
generate_graph()
plt.show()