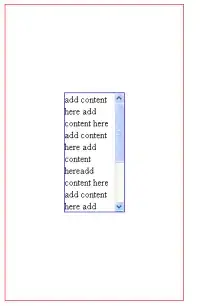
Using the same set of data, I have produced two different tile plots as shown below:
DATA:
> dput(coupler.graph)
structure(list(Category = c("HBC", "TC", "BSC", "GSC", "GSC",
"SSC", "SSC", "GSC", "GSC", "SSC", "SSC", "SSC", "HBC", "TC",
"BSC", "BSC", "GSC", "GSC", "SSC", "HBC", "HBC", "TC", "TC",
"BSC", "GSC", "GSC", "GSC", "GSC", "GSC", "TC", "BSC", "BSC",
"GSC", "GSC"), `Bar Size` = c("No. 5", "No. 5", "No. 5", "No. 5",
"No. 5", "No. 6", "No. 6", "No. 6", "No. 6", "No. 8", "No. 8",
"No. 8", "No. 8", "No. 8", "No. 8", "No. 8", "No. 8", "No. 8",
"No. 10", "No. 10", "No. 10", "No. 10", "No. 10", "No. 10", "No. 10",
"No. 10", "No. 10", "No. 11", "No. 11", "No. 18", "No. 18", "No. 18",
"No. 18", "No. 18"), `No. Bars` = c(3, 9, 3, 6, 6, 85, 85, 7,
7, 90, 90, 90, 7, 9, 6, 6, 21, 21, 9, 22, 22, 27, 27, 13, 25,
25, 25, 8, 8, 4, 4, 4, 4, 4), Failure = c("Bar fracture", "Bar fracture",
"Bar fracture", "Bar pullout", "Bar fracture", "Bar pullout",
"Bar fracture", "Coupler failure", "Bar fracture", "Coupler failure",
"Bar pullout", "Bar fracture", "Bar fracture", "Bar fracture",
"Bar pullout", "Bar fracture", "Bar fracture", "Bar pullout",
"Coupler failure", "Coupler failure", "Bar fracture", "Coupler failure",
"Bar fracture", "Bar fracture", "Bar pullout", "Bar fracture",
"Coupler failure", "Bar fracture", "Coupler failure", "Coupler failure",
"Bar fracture", "Bar pullout", "Bar fracture", "Coupler failure"
), x = c("1-3", "7-9", "1-3", "5-7", "5-7", "30-90", "30-90",
"5-7", "5-7", "30-90", "30-90", "30-90", "5-7", "7-9", "5-7",
"5-7", "20-30", "20-30", "7-9", "20-30", "20-30", "20-30", "20-30",
"11-15", "20-30", "20-30", "20-30", "7-9", "7-9", "3-5", "3-5",
"3-5", "3-5", "3-5")), row.names = c(NA, -34L), class = c("tbl_df",
"tbl", "data.frame"))
The first plot is showing the number of specimens as:
labels1 <- factor(c("0", "1-3", "3-5", "5-7", "7-9",
"9-11", "11-15", "15-20","20-90"), levels =
c("0", "1-3", "3-5", "5-7", "7-9",
"9-11", "11-15", "15-20","20-30","30-90"))
values1 <- c("white", "#ffffd9", "#edf8b1", "#c7e9b4", "#7fcdbb",
"#41b6c4", "#1d91c0", "#225ea8", "#253494","#081d58")
bar_list = c("No. 5", "No. 6", "No. 8", "No. 10", "No. 11", "No. 14", "No. 18")
plot1 <- ggplot(coupler.graph) + aes(x = Category, y = fct_inorder(`Bar Size`), fill = factor(x,
levels = c("0", "1-3", "3-5", "5-7", "7-9",
"9-11", "11-15", "15-20","20-30", "30-90"))) +
geom_tile(width=0.9, height=0.9) + theme_classic() + scale_fill_manual(labels = factor(labels1), values = values1) +
labs(x = "Splicer Type", y = "Bar Size") + scale_y_discrete(limits = bar_list) +
theme(plot.title = element_blank(), axis.text = element_text(color = "black", size = 12), axis.ticks.x = element_blank(),
axis.line = element_line(color = "black", size = 0.2), axis.ticks.y = element_line(color = "black", size = 0.2),
axis.title.y = element_text(color = "black", size = 16, margin = margin(0,40,0,0)),
axis.title.x = element_text(color = "black", size = 16, margin = margin(35,0,0,0)),
legend.title = element_blank(), legend.text = element_text(color = "black", size = 12))
ggplotly(
p = ggplot2::last_plot(),
width = NULL,
height = NULL,
tooltip = c("Category", "Failure"),
dynamicTicks = FALSE,
layerData = 1,
originalData = TRUE,) %>% add_annotations( text="Number of\nSpecimens", xref="paper", yref="paper",
x=1.1, xanchor="left", y=0.8, yanchor="bottom", font = list(size = 18),
legendtitle=TRUE, showarrow=FALSE ) %>%
layout(yaxis = list(title = list(text = "Bar Size", standoff = 30L)),
xaxis = list(title = list(text = "Bar Size",standoff = 30L)),
legend = list(orientation = "v", x = 1.1, y = 0.13))
And the resulted plot is :
The second plot is showing the failure type:
values2 <- c("Bar fracture" = "red", "Bar pullout" = "blue", "Coupler failure" = "yellow")
plot2 <- ggplot(coupler.graph) + aes(x = Category, y = fct_inorder(`Bar Size`), fill = Failure) +
geom_tile(width=0.9, height=0.9) + theme_classic() + scale_fill_manual(values = values2) +
labs(x = "Splicer Type", y = "Bar Size") + scale_y_discrete(limits = bar_list) +
theme(plot.title = element_blank(), axis.text = element_text(color = "black", size = 12), axis.ticks.x = element_blank(),
axis.line = element_line(color = "black", size = 0.2), axis.ticks.y = element_line(color = "black", size = 0.2),
axis.title.y = element_text(color = "black", size = 16, margin = margin(0,40,0,0)),
axis.title.x = element_text(color = "black", size = 16, margin = margin(35,0,0,0)),
legend.title = element_blank(), legend.text = element_text(color = "black", size = 12))
ggplotly(
p = ggplot2::last_plot(),
width = NULL,
height = NULL,
tooltip = c("Category", "Failure"),
dynamicTicks = FALSE,
layerData = 1,
originalData = TRUE,) %>%
layout(yaxis = list(title = list(text = "Bar Size", standoff = 30L)),
xaxis = list(title = list(text = "Bar Size",standoff = 30L)),
legend = list(orientation = "v", x = 1.1, y = 0.13))
And the resulted plot is shown:
I would like to add a button to the HTML output such as the example here does for different chart types, but switching between these two plots.